How Can We Help?
Open Access related release notesOpen Access related release notes
5.30
Mint DOIs for Research Output and related sub-types through DataCite
Pure administrators can now enable minting of DOIs of research output, allowing users to register their research output in DataCite.
It is possible to configure this functionality to allow the minting of DOIs for all research output, or for specific sub-types.

When this functionality is enabled, users have the option to mint the DOI directly from the research output editor. When the DataCite integration is enabled, configured editors and administrators of datasets will be able to mint a new DOI by clicking Create DOI from DataCite.
The Create DOI from DataCite button is only visible to editors and administrators once the record has been saved. It is only possible to mint a DOI at DataCite if no DOI exists on the record.

The following fields are sent and will be registered at DataCite:
- Contributors
- DOI
- Electronic versions
- Journal association
- Original language
- Publication statuses and dates
- Publisher
- Subtitle of the contribution in original language
- Title of the contribution in original language
- Translated subtitle of the contribution
- Translated title of the contribution
Once a DOI has been minted these fields will be locked for the personal user. If the metadata on the record is updated by the editor or administrator, the update is sent to DataCite and the record will be updated.
It is not possible to change the ‘mandatory’ metadata on that record. If the metadata is updated, a new DOI must be minted.
Note: It is not possible to mint DOIs on the following:
-
Research output where the visibility is ‘backend’ or ‘confidential’
-
Research output that has been imported to Pure through an online source
Requirements
The integration with DataCite is only available to customers who have a Pure portal and is disabled by default.
To use this integration you must first register with DataCite. Contact them at contact@datacite.org and request the necessary access.
For more information, see the Metadata Store website at https://mds.datacite.org.
Once you have received the credentials, administrators can configure the minting of DOIs from DataCite for research output and datasets:
- Administrator > Integrations > DataCite DOI Minting
Available resources
Related release notes:
- https://helpcenter.pure.elsevier.com/open-access/registering-datasets-at-datacite
- https://helpcenter.pure.elsevier.com/en_US/51/release-notes-pure-510-4200#updated-datasets-model-and-integration-to-datacite-metadata-store-3
5.29
Funding information import settings
Added control for funding information in Pure
Pure administrators now have more fine-grained control on how funding information including funding acknowledgement texts and funding details such as funding organisations and numbers are imported with research outputs and datasets. The import controls limit what is imported by either users or jobs. These settings do not affect any funding information-related rules created in Available updates.
From 5.29.2, funding information is by default NOT imported.
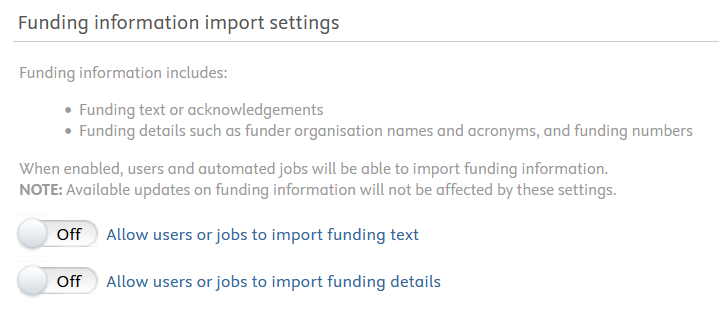
Requirements
For RESEARCH OUTPUS, to enable import of funding information by users and jobs, go to Administrator > Research output > Import configuration > Funding information import settings.
For DATASETS, to enable import of funding information by users and jobs, go to Administrator > Datasets > Configuration > Funding information import settings.
5.28
Funding information on RO and Datasets
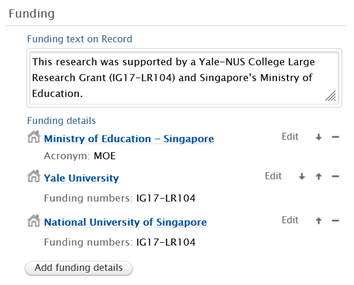
Dedicated funding information section on Research output and Dataset editors
Funding information, including funding acknowledgement text and abbreviated funder and funding number details, are available in a dedicated funding section on research output and dataset records. External harvesting services utilizing e.g. OAI-PMH or Pure API endpoints, are able to access the full acknowledgement text.
Funding information from Research outputs were previously captured in the bibliographic note section of the Research output editor and will now only be available in the funding section.
Funding text and abbreviated funding details can be manually added to a record, and funding information from Research outputs will be added automatically when imported from Scopus or Web of Science (WoS). Funding information updates are also possible via the Available updates functionality.
Info
The funding section is immediately available and does not require any additional actions to enable or display the information.
On update to 5.28.0, a dedicated backfill job will automatically scan all research output XML files and add any funding acknowledgement texts to the new funding section on the editor. No configuration is required on the part of a Pure administrator.
Info
The funding section on Research outputs and Datasets may duplicate information found in related Project and Award records. We recommend that institution-relevant funding details always be captured on awards and related to research outputs and datasets via projects.
For more information and guidance on how to configure available updates to process funding acknowledgement text, please review the Pure 5.13 release notes
For more information on related OAI-PMH updates in this release Funding on datasets and pulbications in OAI-PMH.
Read more about how Pure supports Open Access and compliance.
Pure Manual > Pure for Technical Administrators > Core functionality > Content import, synchronisation, and export > Open Access support in Pure
OAI-PMH: Datasets
In line with the Plan S requirements for Open Access compliance, in this release we have expanded support for OAI-PMH in Pure.
It is now possible to export datasets via the standard OAI-PMH metadata formats: oai_cerif_openaire (CERIF), mods (MODS), and oai_dc (DC).
The following new fields are available on datasets in CERIF:
- Funding information
- Dates
- File locations
Datasets are included also in MODS and Dublin Core.
You can list all datasets via the new OAI-PMH set: datasets:all
<cerif:OriginatesFrom>
<cerif:Funding>
<funt:Type>https://www.openaire.eu/cerif-profile/vocab/OpenAIRE_Funding_Types#Grant</funt:Type>
<cerif:Acronym>TUG</cerif:Acronym>
<cerif:Identifier type="/dk/atira/funding/fundingdetails/fundingnumber">TUGU-5678</cerif:Identifier>
<cerif:Description xml:lang="en">This work was funded by Test University and Another University.</cerif:Description>
<cerif:Funder>
<cerif:OrgUnit id="75cbdb07-a7b5-4504-8623-b4481c1b68b6">
<cerif:Name xml:lang="en">Test University</cerif:Name>
</cerif:OrgUnit>
</cerif:Funder>
</cerif:Funding>
</cerif:OriginatesFrom>
<cerif:OriginatesFrom>
<cerif:Funding>
<funt:Type>https://www.openaire.eu/cerif-profile/vocab/OpenAIRE_Funding_Types#Grant</funt:Type>
<cerif:Description xml:lang="en">This work was funded by Test University and Another University.</cerif:Description>
<cerif:Funder>
<cerif:OrgUnit id="0630eb3b-1c01-42b1-aba9-ccbcb059844f">
<cerif:Name xml:lang="en">Another University</cerif:Name>
</cerif:OrgUnit>
</cerif:Funder>
</cerif:Funding>
</cerif:OriginatesFrom> |
OAI-PMH: Funding information
As part of expanding support for OAI-PMH we have included funding information.
All funding information is now available via OAI-PMH in the CERIF format, while MODS includes only the funding text.
<cerif:OriginatesFrom>
<cerif:Funding>
<funt:Type>https://www.openaire.eu/cerif-profile/vocab/OpenAIRE_Funding_Types#Grant</funt:Type>
<cerif:Acronym>NSFC</cerif:Acronym>
<cerif:Identifier type="/dk/atira/funding/fundingdetails/fundingnumber">822044593</cerif:Identifier>
<cerif:Description xml:lang="en">This study is funded by the National Natural Science Foundation of China (No. 822044593 ), the State Key Laboratory for Chemistry and Molecular Engineering of Medicinal Resources ( Guangxi Normal University ) ( CMEMR2022-B11 ), and the Natural Science Foundation of the Jiangsu Higher Education Institutions of China ( 22KJB360018 and 22KJB310025 ).</cerif:Description>
<cerif:Funder>
<cerif:OrgUnit id="3ed69b17-00f3-4976-baa9-097b31aa10d5">
<cerif:Name xml:lang="en">National Natural Science Foundation of China</cerif:Name>
</cerif:OrgUnit>
</cerif:Funder>
</cerif:Funding>
</cerif:OriginatesFrom>
<cerif:OriginatesFrom>
<cerif:Funding>
<funt:Type>https://www.openaire.eu/cerif-profile/vocab/OpenAIRE_Funding_Types#Grant</funt:Type>
<cerif:Acronym>GXNU</cerif:Acronym>
<cerif:Identifier type="/dk/atira/funding/fundingdetails/fundingnumber">CMEMR2022-B11</cerif:Identifier>
<cerif:Description xml:lang="en">This study is funded by the National Natural Science Foundation of China (No. 822044593 ), the State Key Laboratory for Chemistry and Molecular Engineering of Medicinal Resources ( Guangxi Normal University ) ( CMEMR2022-B11 ), and the Natural Science Foundation of the Jiangsu Higher Education Institutions of China ( 22KJB360018 and 22KJB310025 ).</cerif:Description>
<cerif:Funder>
<cerif:OrgUnit id="c6db9909-a466-4338-ac11-b4f2cb7ee264">
<cerif:Name xml:lang="en">Guangxi Normal University</cerif:Name>
</cerif:OrgUnit>
</cerif:Funder>
</cerif:Funding>
</cerif:OriginatesFrom>
<cerif:OriginatesFrom>
<cerif:Funding>
<funt:Type>https://www.openaire.eu/cerif-profile/vocab/OpenAIRE_Funding_Types#Grant</funt:Type>
<cerif:Identifier type="/dk/atira/funding/fundingdetails/fundingnumber">22KJB310025</cerif:Identifier>
<cerif:Identifier type="/dk/atira/funding/fundingdetails/fundingnumber">22KJB360018</cerif:Identifier>
<cerif:Description xml:lang="en">This study is funded by the National Natural Science Foundation of China (No. 822044593 ), the State Key Laboratory for Chemistry and Molecular Engineering of Medicinal Resources ( Guangxi Normal University ) ( CMEMR2022-B11 ), and the Natural Science Foundation of the Jiangsu Higher Education Institutions of China ( 22KJB360018 and 22KJB310025 ).</cerif:Description>
<cerif:Funder>
<cerif:OrgUnit id="e99ea5a8-4ad0-4a98-9924-06fb7c60bab2">
<cerif:Name xml:lang="en">Natural Science Research of Jiangsu Higher Education Institutions of China</cerif:Name>
</cerif:OrgUnit>
</cerif:Funder>
</cerif:Funding>
</cerif:OriginatesFrom>
<cerif:OriginatesFrom>
<cerif:Funding>
<funt:Type>https://www.openaire.eu/cerif-profile/vocab/OpenAIRE_Funding_Types#Grant</funt:Type>
<cerif:Description xml:lang="en">This study is funded by the National Natural Science Foundation of China (No. 822044593 ), the State Key Laboratory for Chemistry and Molecular Engineering of Medicinal Resources ( Guangxi Normal University ) ( CMEMR2022-B11 ), and the Natural Science Foundation of the Jiangsu Higher Education Institutions of China ( 22KJB360018 and 22KJB310025 ).</cerif:Description>
<cerif:Funder>
<cerif:OrgUnit id="c9f3631e-fc9f-4130-8287-518409a8114c">
<cerif:Name xml:lang="en">Key Laboratory of Bioorganic Chemistry and Molecular Engineering, Ministry of Education</cerif:Name>
</cerif:OrgUnit>
</cerif:Funder>
</cerif:Funding>
</cerif:OriginatesFrom> |
CERIF
Funding details is now included in CERIF for both research output, patents, and datasets. An example of this output is shown below.
MODS
Funding text is now included in MODS for both research output, patents, and datasets. An example of this output is shown below.
<mods:note type="funding" xlink:type="simple" xml:lang="eng">This study is funded by the National Natural Science Foundation of China (No. 822044593 ), the State Key Laboratory for Chemistry and Molecular Engineering of Medicinal Resources ( Guangxi Normal University ) ( CMEMR2022-B11 ), and the Natural Science Foundation of the Jiangsu Higher Education Institutions of China ( 22KJB360018 and 22KJB310025 ).</mods:note> |
DC
Dublin Core does not have any suitable fields for funding information.
OAI-PMH: Files on Research Output and Datasets
In release 5.28 of Pure we have expanded the details available on electronic version files on research outputs and of files on datasets. These are available in the standard formats: CERIF, MODS, and Dublin Core.
CERIF
Electronic version files are now available in CERIF, as shown in the example below:
<cerif:FileLocations>
<cerif:Medium>
<cerif:Type scheme="/dk/atira/pure/researchoutput/electronicversion/versiontype">publishersversion</cerif:Type>
<cerif:Title xml:lang="en">report.pdf</cerif:Title>
<cerif:URI>http://<pure-server>/portal/files/25927/report.pdf</cerif:URI>
<cerif:MimeType>application/pdf</cerif:MimeType>
<cerif:Size>20043</cerif:Size>
<ar:Access>http://purl.org/coar/access_right/c_abf2</ar:Access>
<cerif:License scheme="/dk/atira/pure/core/document/licenses">cc_by</cerif:License>
</cerif:Medium>
</cerif:FileLocations> |
MODS
In MODS, electronic version files are now expressed in a single element (physicalDescription). File size and individual access status is now available.
<mods:note type="version identification" displayLabel="report.pdf" ID="file_20701">publishersversion</mods:note> <mods:note type="license" displayLabel="report.pdf" ID="file_20701">cc_by</mods:note> <mods:note type="document visibility" displayLabel="report.pdf" ID="file_20701">FREE</mods:note> |
<mods:physicalDescription> <mods:form authority="marcform">electronic</mods:form> <mods:note type="title" xlink:type="simple">report</mods:note> <mods:internetMediaType>application/pdf</mods:internetMediaType> <mods:extent unit="bytes">20043</mods:extent> <mods:note type="location" xlink:type="simple" xlink:href="http://<pure-server>/portal/files/25927/report.pdf"/> <mods:note type="visibility" xlink:type="simple">FREE</mods:note> <mods:note type="license" typeURI="/dk/atira/pure/core/document/licenses" xlink:type="simple">cc_by</mods:note> <mods:note type="version" typeURI="/dk/atira/pure/researchoutput/electronicversion/versiontype" xlink:type="simple">publishersversion</mods:note> <mods:note type="access" typeURI="/dk/atira/pure/core/openaccesspermission" xlink:type="simple">open</mods:note> </mods:physicalDescription> |
Dublin Core
The export of electronic files is now also included in Dublin Core.
<identifier>https://<pure-server>/ws/files/20702/report.pdf</identifier> |
OAI-PMH: Identifier types and name identifiers
We have improved support for identifiers on research output and datasets in MODS and Dublin Core, such that the identifier type and url are shown together with the ID.
MODS
Name identifiers and identifier types/URIs has been added to MODS:
<mods:identifier type="local">PURE: 19412</mods:identifier> <mods:identifier type="local">PURE UUID: 5fb04e8f-2f48-4942-bb63-1df82f9aa63a</mods:identifier> <mods:identifier type="local">Scopus: 85168535696</mods:identifier> <mods:identifier type="local">PubMed: 37597675</mods:identifier> <mods:identifier type="local">handle.net: https://hdl.handle.net/test_university_pure/5fb04e8f-2f48-4942-bb63-1df82f9aa63a</mods:identifier> <mods:identifier type="doi">https://doi.org/10.1016/j.jep.2023.117058</mods:identifier> |
<mods:identifier type="pure/id">25923</mods:identifier> <mods:identifier type="pure/uuid">1ca877f1-4b75-44d5-a003-13e246dff730</mods:identifier> <mods:identifier type="scopus">85168535696</mods:identifier> <mods:identifier type="pmid">37597675</mods:identifier> <mods:identifier type="hdl" typeURI="http://id.loc.gov/vocabulary/identifiers/hdl">test_university_pure/5fb04e8f-2f48-4942-bb63-1df82f9aa63a</mods:identifier> <mods:identifier type="doi" typeURI="http://id.loc.gov/vocabulary/identifiers/doi">10.1016/j.jep.2023.117058</mods:identifier> |
MODS: NAME IDENTIFIERS (NEW)
<mods:name type="personal" xlink:type="simple" authority="local">
<mods:role>
<mods:roleTerm type="text" authority="pure/role">author</mods:roleTerm>
</mods:role>
<mods:nameIdentifier type="pure/uuid">d5ef5eea-e0a0-4e84-bb16-75fd251eb1db</mods:nameIdentifier>
<mods:nameIdentifier type="isni" typeURI="http://id.loc.gov/vocabulary/identifiers/isni">000000022265270X</mods:nameIdentifier>
<mods:nameIdentifier type="dai-nl" typeURI="info:eu-repo/dai/nl">30435354X</mods:nameIdentifier>
<mods:nameIdentifier type="orcid" typeURI="http://id.loc.gov/vocabulary/identifiers/orcid">0000-0002-2265-270X</mods:nameIdentifier>
<mods:namePart type="given">John 'Internal-Person'</mods:namePart>
<mods:namePart type="family">Doe</mods:namePart>
<mods:affiliation>Test University</mods:affiliation>
</mods:name> |
Dublin Core
Identifiers in DC now have types:
<identifier>https://doi.org/10.1016/j.jep.2023.117058</identifier> <identifier>https://hdl.handle.net/test_university_pure/5fb04e8f-2f48-4942-bb63-1df82f9aa63a</identifier> <identifier>https://<pure-server>/ws/files/20702/report.pdf</identifier> <identifier>http://www.scopus.com/inward/record.url?scp=85168535696&partnerID=8YFLogxK</identifier> |
<dc:identifier type="doi">https://doi.org/10.1016/j.jep.2023.117058</dc:identifier> <dc:identifier type="handle.net">https://hdl.handle.net/test_university_pure/5fb04e8f-2f48-4942-bb63-1df82f9aa63a</dc:identifier> <dc:identifier type="/dk/atira/pure/researchoutput/electronicversion/versiontype/publishersversion">https://<pure-server>/ws/files/19710/report.pdf</dc:identifier> <dc:identifier type="/dk/atira/pure/links/researchoutput">http://www.scopus.com/inward/record.url?scp=85168535696&partnerID=8YFLogxK</dc:identifier> |
OAI-PMH: Schema upgrades
In this release we have also updated the metadata schemas for CERIF and MODS. the CERIF schema has been upgraded to version 1.2 and the MODS schema has been updated to version 3.8. There is no change to the Dublin Core metadata schema.
More details on these updates are listed below.
CERIF (1.1 → 1.2)
We have upgraded to Openaire CERIF Profile schema version 1.2 (released June 2023).
More: https://openaire-guidelines-for-cris-managers.readthedocs.io/en/v1.2.0/introduction.html#versions
Note on CERIF XML import backwards compatibility: Pure is not able to import old CERIF XML exports (v1.1).
MODS (3.3 → 3.8)
We have upgraded to MODS schema version 3.8 (released September 2022).
More: https://loc.gov/standards/mods/mods-3-8-announcement.html
DC (1.1 – no update)
We are already on the latest DCMES schema version 1.1 (released February 2008).
OAI-PMH: Miscellaneous
In this section we describe additional updates to OAI-PMH that are included in release 5.28.
CERIF
We have corrected the way we output classifications.
<cerif:Status scheme="/dk/atira/pure/researchoutput/status/epub">E-pub ahead of print</cerif:Status> |
MODS
Preprint template type is now visible in MODS.
<mods:genre authority="dct" type="publicationType">Preprint</mods:genre> |
DC
Dublin Core elements now use the dc: namespace prefix.
Keywords are exported in all available languages, and classified keywords have been grouped:
<subject>Chemical interaction</subject> <subject>Combination mechanism</subject> <subject>Ischemic stroke</subject> <subject>Ligusticum chuanxiong hort</subject> <subject>Salvia miltiorrhiza bunge</subject> <subject>Synergistic effect</subject> <subject>/dk/atira/pure/subjectarea/asjc/3000/3004</subject> <subject>Pharmacology</subject> <subject>/dk/atira/pure/subjectarea/asjc/3000/3002</subject> <subject>Drug Discovery</subject> |
<dc:subject xml:lang="eng">Chemical interaction</dc:subject> <dc:subject xml:lang="eng">Combination mechanism</dc:subject> <dc:subject xml:lang="eng">Ischemic stroke</dc:subject> <dc:subject xml:lang="eng">Ligusticum chuanxiong hort</dc:subject> <dc:subject xml:lang="eng">Salvia miltiorrhiza bunge</dc:subject> <dc:subject xml:lang="eng">Synergistic effect</dc:subject> <dc:subject>/dk/atira/pure/subjectarea/asjc/3000/3004; name=Pharmacology</dc:subject> <dc:subject>/dk/atira/pure/subjectarea/asjc/3000/3002; name=Drug Discovery</dc:subject> |
Pure API: Datasets and equipment/facilities support
The Pure API now supports the management of datasets and equipment/facilities.
5.25
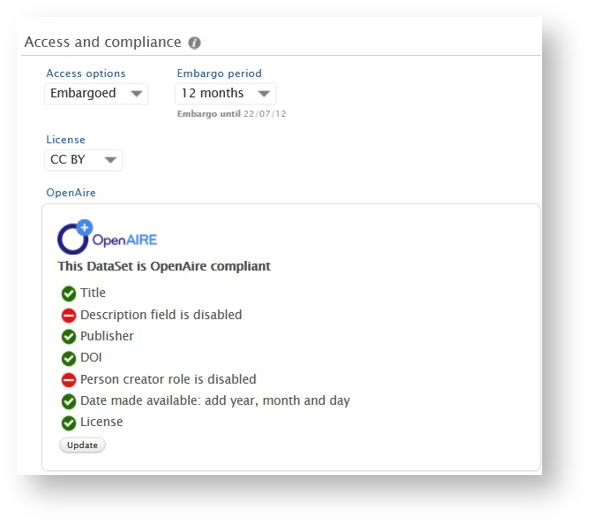
Datasets: OpenAIRE-required fields and status check on datasets
In line with Pure's 5Cs data quality framework, we have expanded the model for dataset records in Pure to align with OpenAIRE guidelines for dataset interoperability (https://guidelines.openaire.eu/en/latest/data/index.html). To facilitate the enrichment, monitoring and management of datasets for OA compliance, including all mandatory OpenAIRE fields, it is now possible for users to add License information at the record and file level. The users can also get an easy overview of whether a particular dataset is OpenAIRE-compliant through the newly added OpenAIRE compliance check.
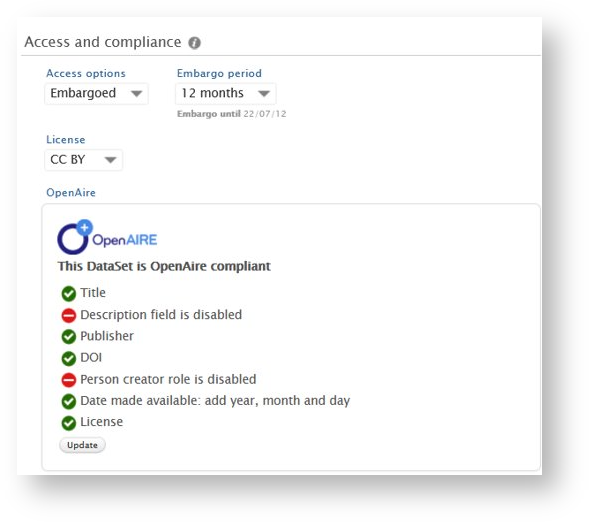
License information on a record
Users can specify a data-specific license on the dataset, with values provided in the modifiable classification 'Datasets: Document licenses' (
License information on a record
Users can specify a data-specific license on the dataset, with values provided in the modifiable classification 'Datasets: Document licenses' (
-
- If there is a license set on the dataset, this will now be exposed as part of product set (see CERIF XML mapping for more details).
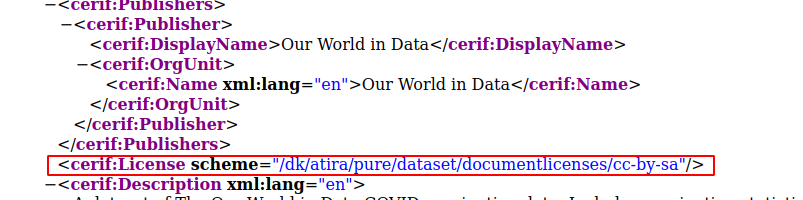
-
- The configurable dataset synchronization now supports synchronizing the license information if data is provided for it in the XML.
- The dataset bulk import functionality now supports importing in the license information if data is provided for it in the XML.
-
|
|
|
Updating values in some fields will not trigger immediate updates to the compliance check, but users can click the Update button to trigger an update.
|
|
Filters and availability of compliance status in reporting
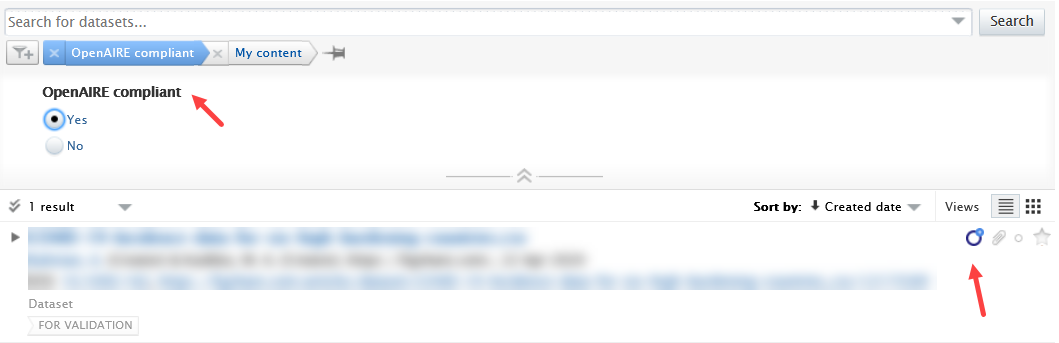
The OpenAIRE compliance status can be used elsewhere in Pure to help identify records that need enrichment.
|
OpenAIRE status filter When the compliance check is enabled, a corresponding filter in the dataset overview will become active, and each compliant record will be tagged with an OpenAIRE icon. |
|
|
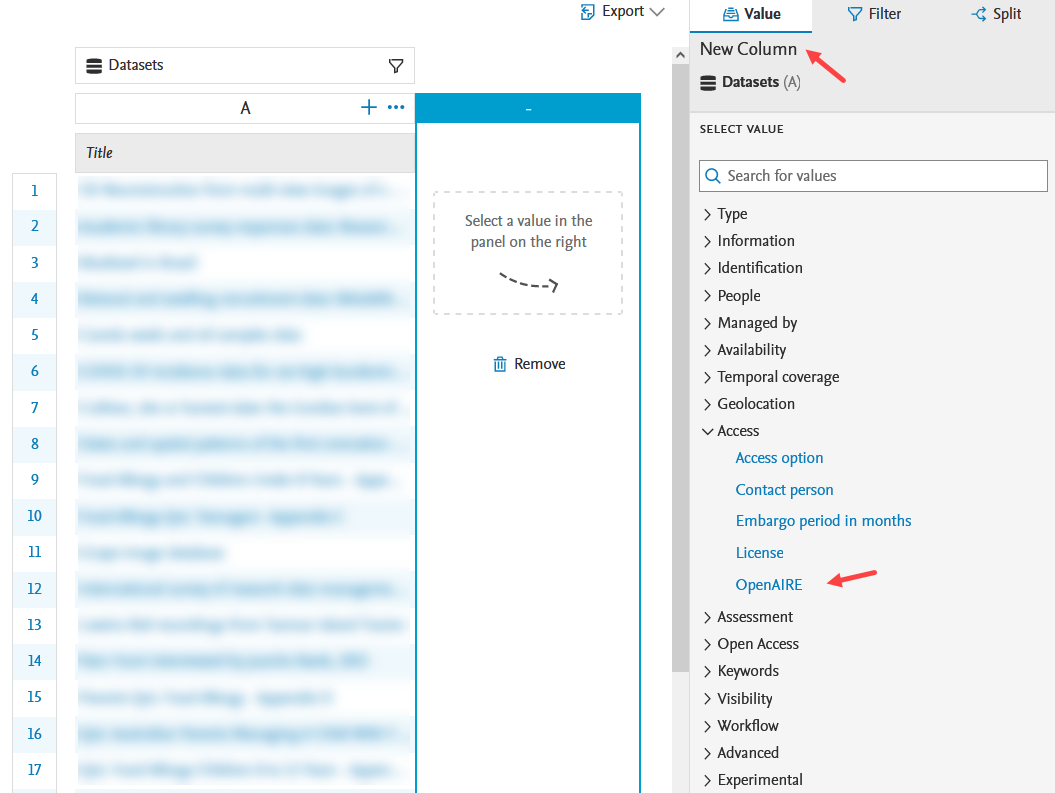
OpenAIRE status as column value in reporting The compliance status can be used as a column value in reporting. It can be found under the Access section. |
|
|
OpenAIRE status as filter in reporting Compliance status can also be used as a filter in reporting. |
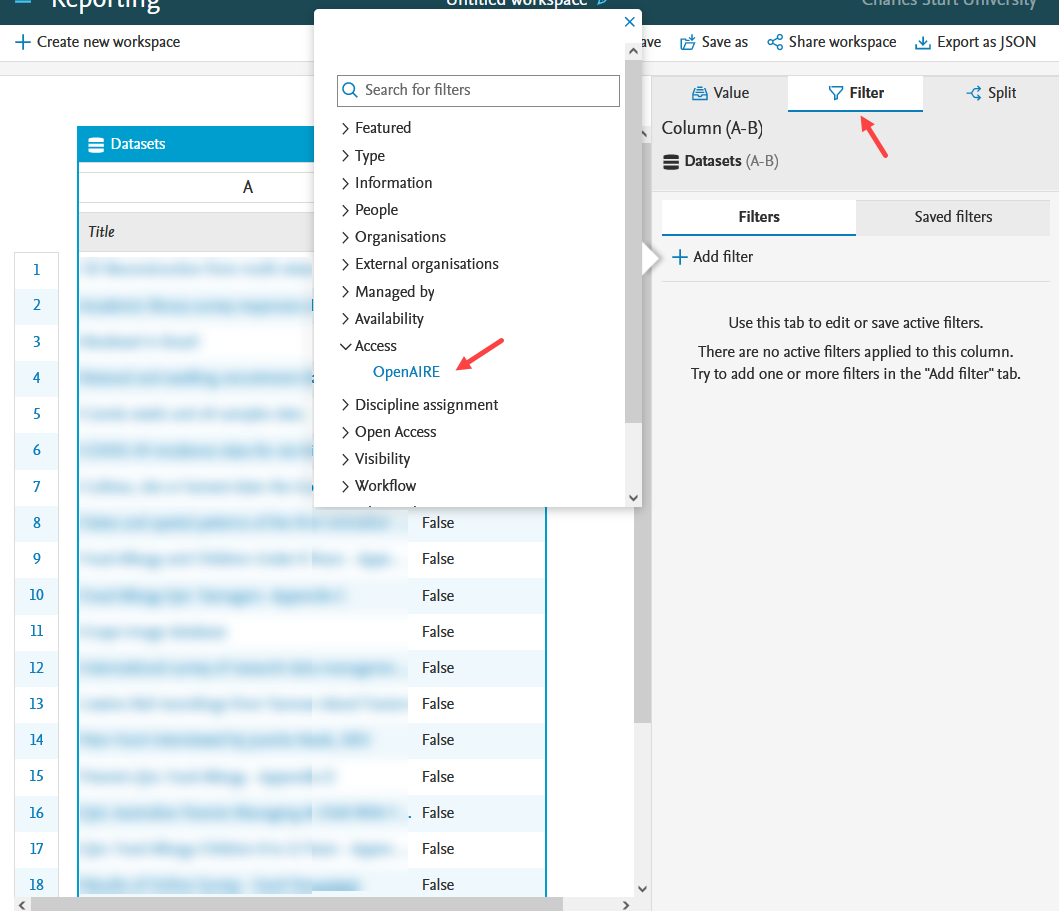
|
5.21
Introducing the preprint sub-type
A user can now import, create and report on preprints in much the same way they can with other research outputs.
In support of Open Science and Open Access, preprints are now an integral part of Pure. This gives researchers more options to disseminate their research outputs and outcomes. A preprint is typically defined as a complete written description of a body of scientific work that has yet to be published in a journal. A preprint can also function as a commentary, a report of negative results, a large data set and its description, and more. It can also be a paper that has been peer-reviewed and is either awaiting formal publication by a journal, or was rejected but the authors are willing to make the content public. In short, a preprint is a research output that has not completed a typical publication pipeline but is of value to the community and deserving of being easily discovered and accessed.
Enabling preprint sub-type
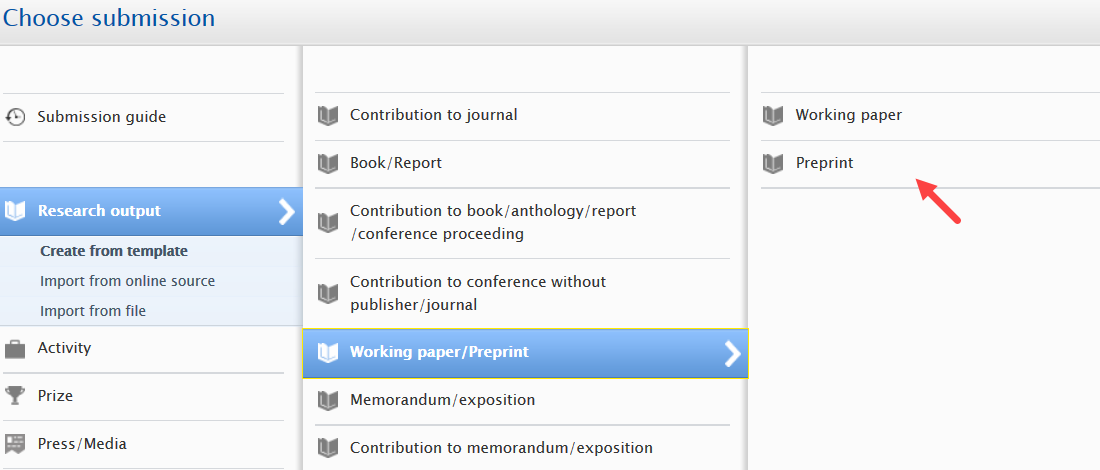
|
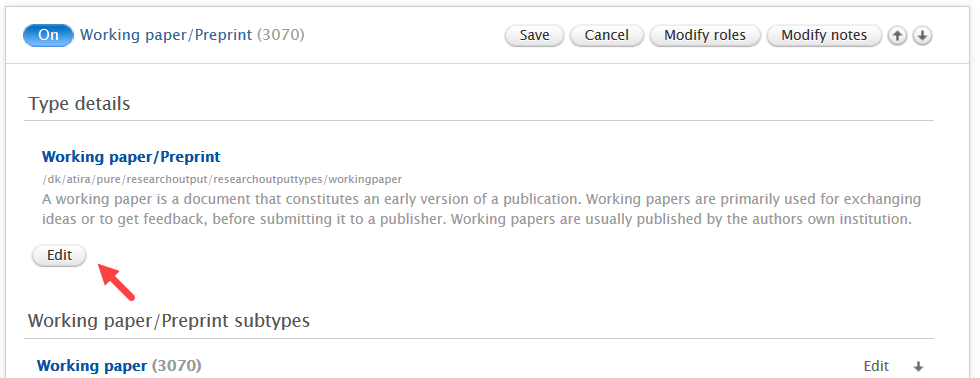
Administrators can enable the preprint sub-type in Administrator > Research outputs > Types:
|
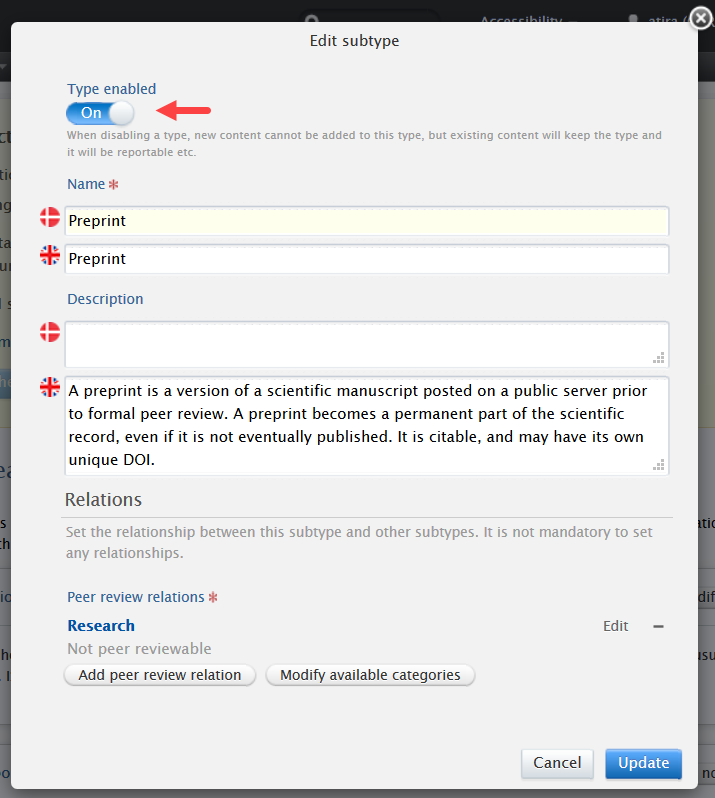
|
We suggest Administrators update the name of 'Working paper' template to 'Working paper/Preprint' so that users can find the Preprint sub-type more easily. To do so, edit the 'Working paper' template and update the name.
We also recommend description updates if your institution has preferred terminologies for working papers and preprints.
Always remember to click Update and Save after making any changes to a template.
|
|
Selecting Preprint sub-type as a user
When enabled, Preprints are displayed as a sub-type of the working paper template. To manually create a preprint, users should select 'Research output' - 'Working paper/Preprint' template and then select the 'Preprint' sub-type.

Importing preprints
Specific import sources have been mapped to the preprint sub-type. When a user imports content from:
- ArXiv
- MedRxiv
- BioRxiv or
- SSRN
The import will appear by default as a preprint. Users will still be able to change templates if necessary.
When importing from other research output sources not listed above, if no journal data is found on the record, the imported content will also be mapped to the preprint sub-type.
The preprint editor
The fields available on the preprint editor are as comprehensive as possible to cater for different institutions needs and interpretations of what a preprint can be.
We highly recommend adding your users' most frequently used preprint servers as Publishers to help them maintain the best possible record.
Migrating content to the Preprint sub-type
If your Pure already has preprints (but defined under different research output templates/sub-types), you can bulk change these to use the preprints sub-type. However, please note that some fields (such as Journal data in a Journal Article) may not be mapped to the preprint sub-type. Any fields not mapped will be added in the history and comments tab of the specific content.
Preprints in Reporting and Web Service
As the new preprints sub-type is part of the working paper template, it is automatically available in reporting and the WS.
5.20
New Import Source: medRxiv
medRxiv (https://www.medrxiv.org/) is a (Open) preprint service for the Medical, Clinical and related Health Sciences.
The integration with medRxiv provides researchers and institution with content in Medicine and Health Sciences, especially relevant at this time to stay up-to-date with the newest COVID-related studies and results.
Instructions on how to enable, add, and search for content from medRxiv are shown below.
| Instructions | Screenshot |
|---|---|
| To enable medRxiv as an import source, go to Administrator > Research Output > Import Sources. | 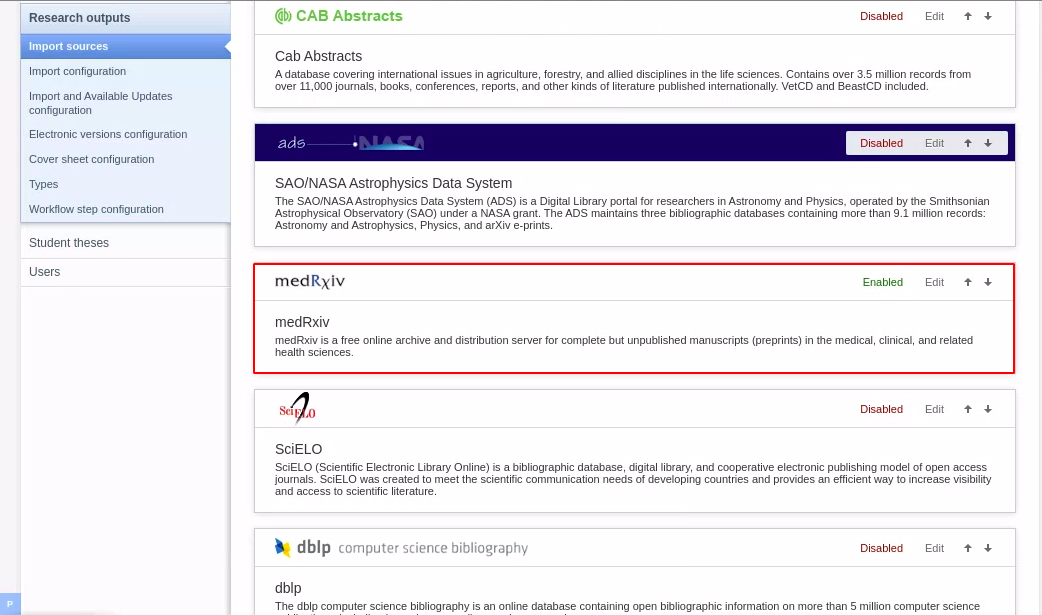
|
| To search for and import content, go to Editor > Research Output > Import from online sources and select medRxiv. | 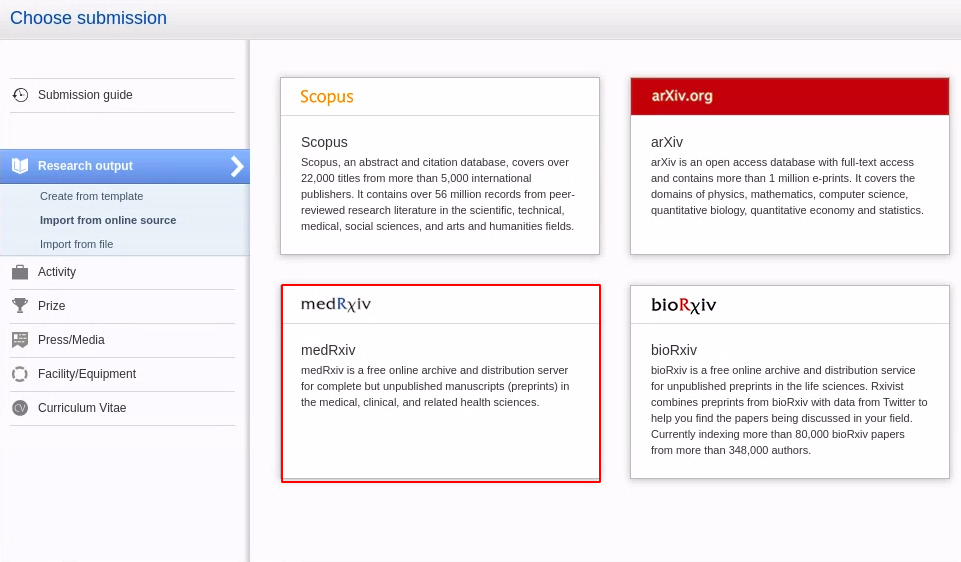
|
|
There is one option when searching for content:
Search results are presented as import candidates and can then be previewed and imported (or removed). Records that already exist in Pure are shown as duplicates. |
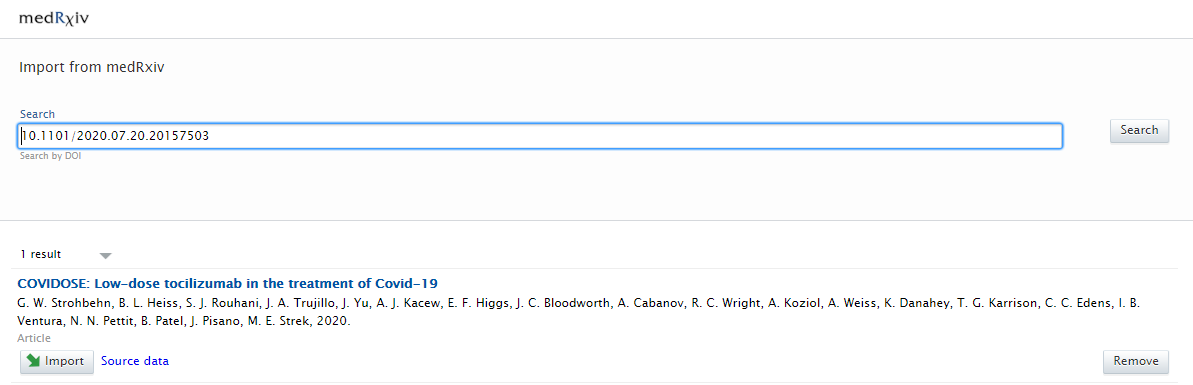
|
Self-import for datasets
In this release we are happy to announce that we expanded functionality for datasets in Pure, making it possible for researchers and personal users to search for and import datasets from specific repositories.
In addition, Mendeley Data Monitor has been added as a self-import source, allowing scientists and researchers to search for datasets across a variety of domain-specific and cross-domain institutional repositories and other data sources. Researchers can quickly preview and assess datasets before accessing them in the destination repository.
Instructions on how to enable, add and search for content from Mendeley Data Monitor are shown below.
| Instructions | Screenshot |
|---|---|
| To enable Data Monitor as an import source, go to Administrator > Datasets > Import Sources. | 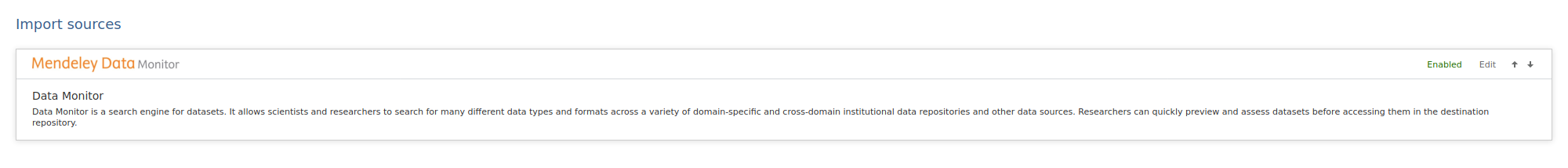
|
|
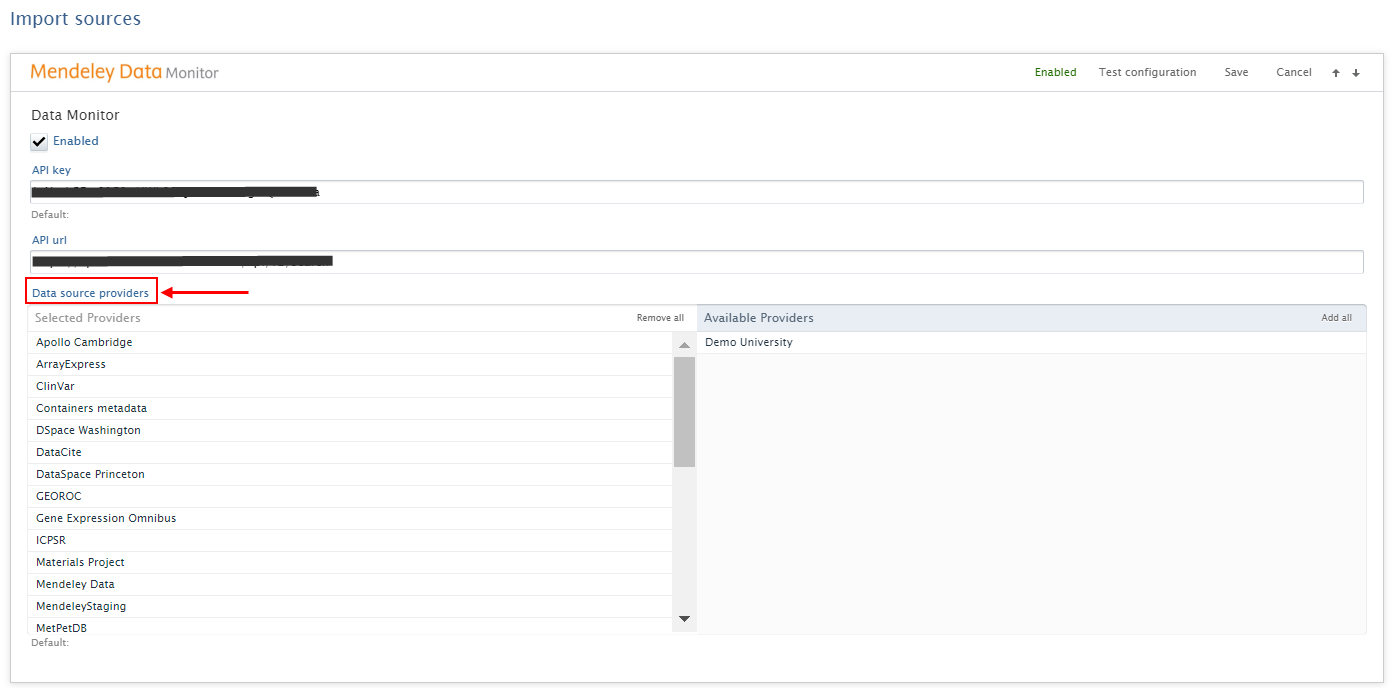
In the source configuration it is possible to insert the API key and a URL. It is possible to enable Mendeley Data Monitor as an import source with or without an API key. Available data sources can then be selected under Data Source Providers. Sources listed on the right will be excluded from the search. After setting the desired configuration, click Save. |
|
| To search and import content, go to Editor > Dataset > Import from online sources and select Mendeley Data Monitor. | 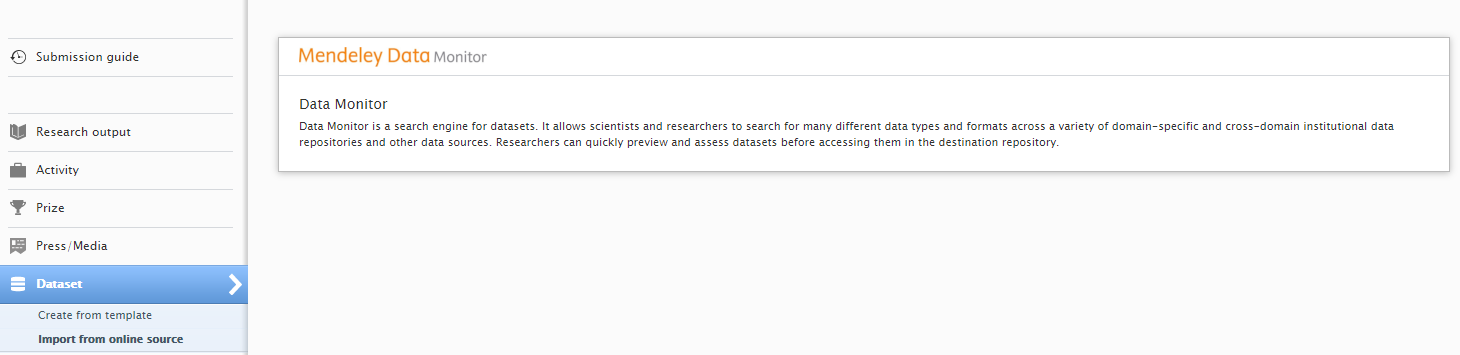
|
|
You can now search content using query text, title, author name(s), DOI, and/or publication year. You can also combine the values. For instance, if you are interested in content with a certain title and author from a specific year, then you can fill out all of the fields and do a combined search. The query field is a text field where you can type out your search criteria. You can find possible search queries here: https://data.mendeley.com/faq?q=advanced+search#data-search-data-search-how-can-i-advanced |
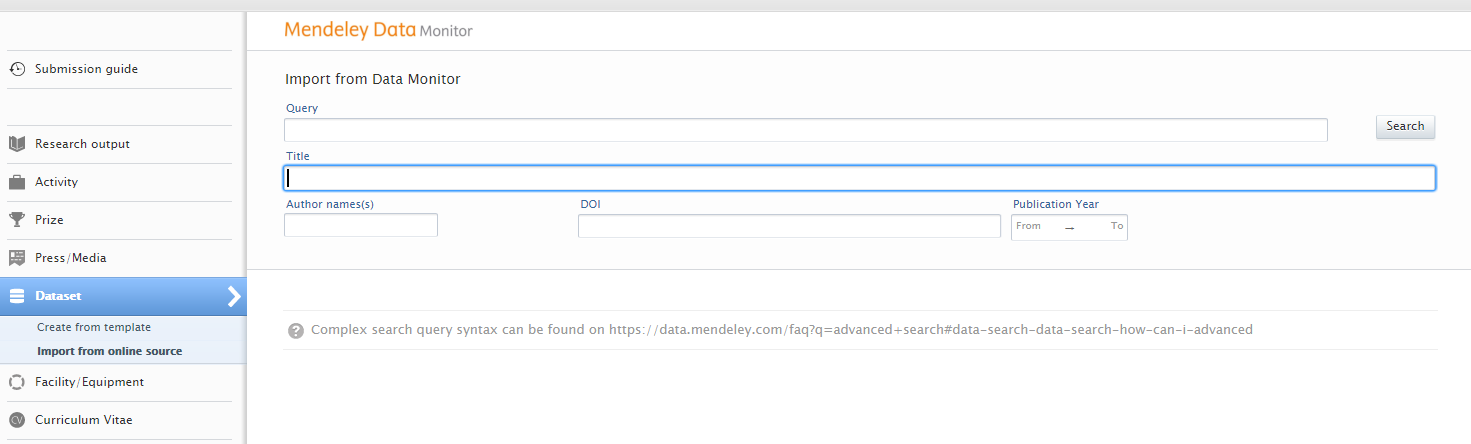 |
|
Search results are presented as import candidates and can then be previewed and imported (or removed). Datasets that have previously been imported to Pure are shown as duplicates. The data source from which the dataset is being imported is shown for all import candidates. |
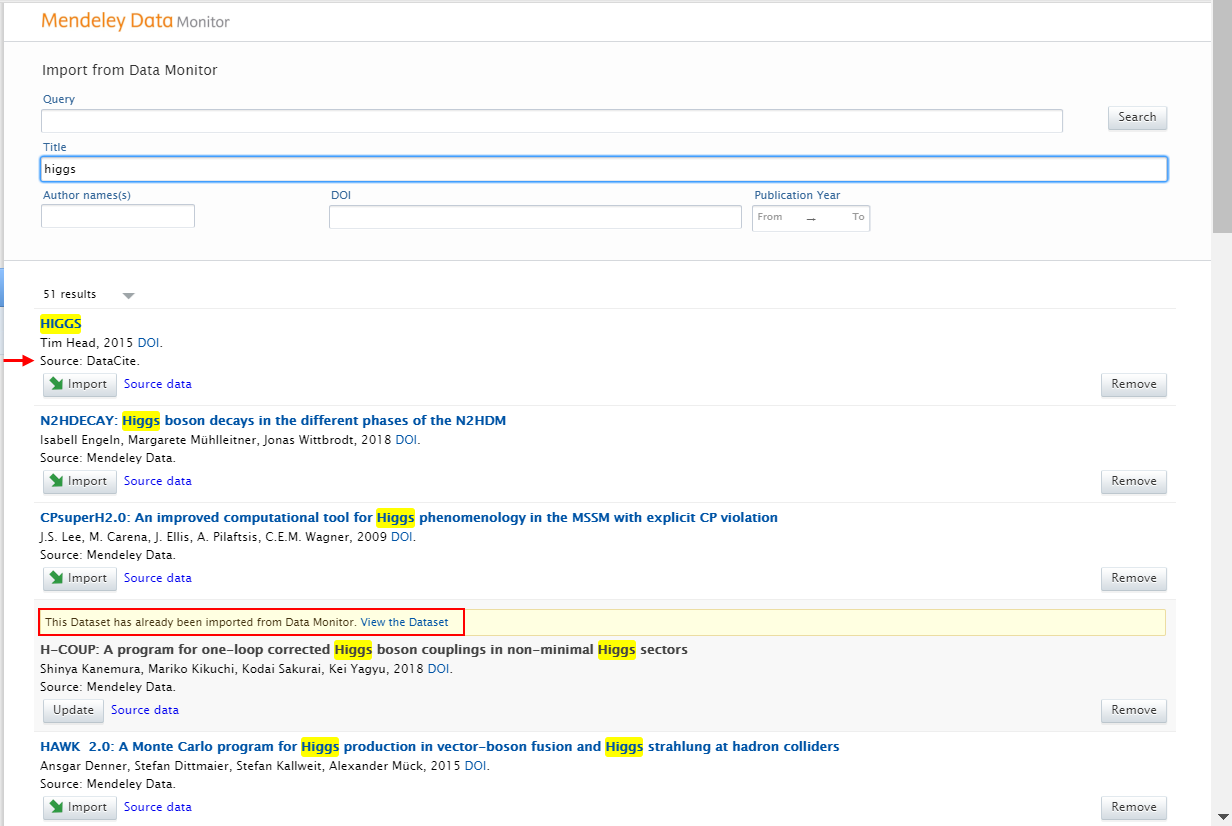
|
Reporting on Open access
With this release, we have added more values and filters on Open Access and APC . This will make it possible to create reports on Research outputs and their Open Access status, embargo date, APC information, and more.
We are currently working on providing standard reports that make use of the new values and filters. We hope to make these available with the 5.20.1 release so please an eye on the future release notes.
We have added more filters on Open Access. Use these to limit the content you want to include in your report.
These are filters on the open access status of the research output.
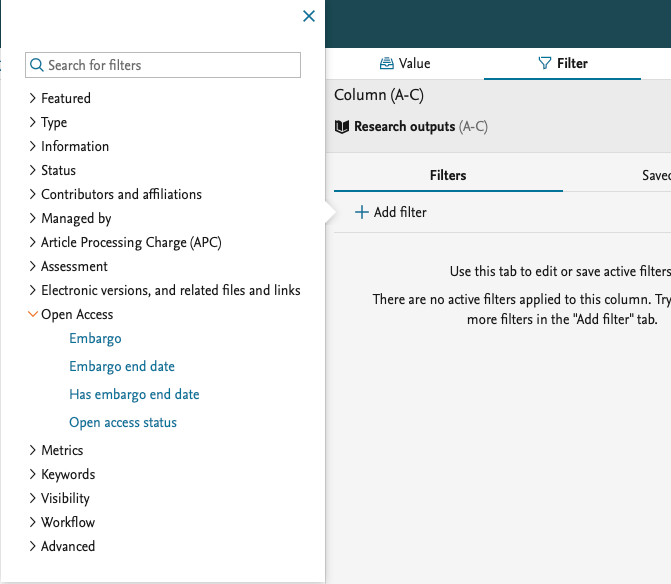
We have also added more filters on the electronic versions on the Research output, which allow you to limit the report to content that has an Open Access document added
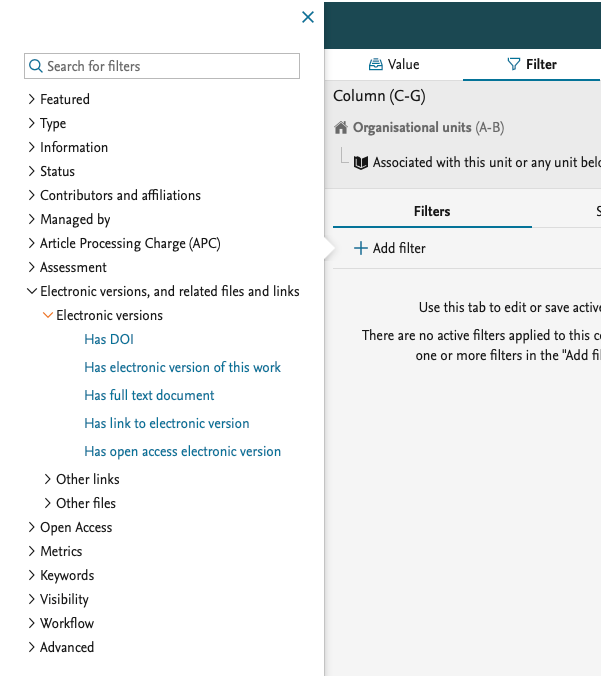
We have added more values that can be used to create the data you need for your report. You can now project the 'Embargo end date' and the overall 'Open Access status' of the Research output.
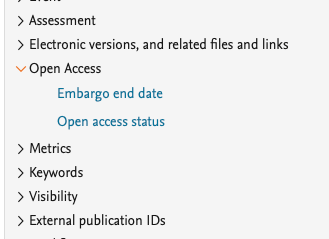
We have updated the ability to split data by Open Access status:
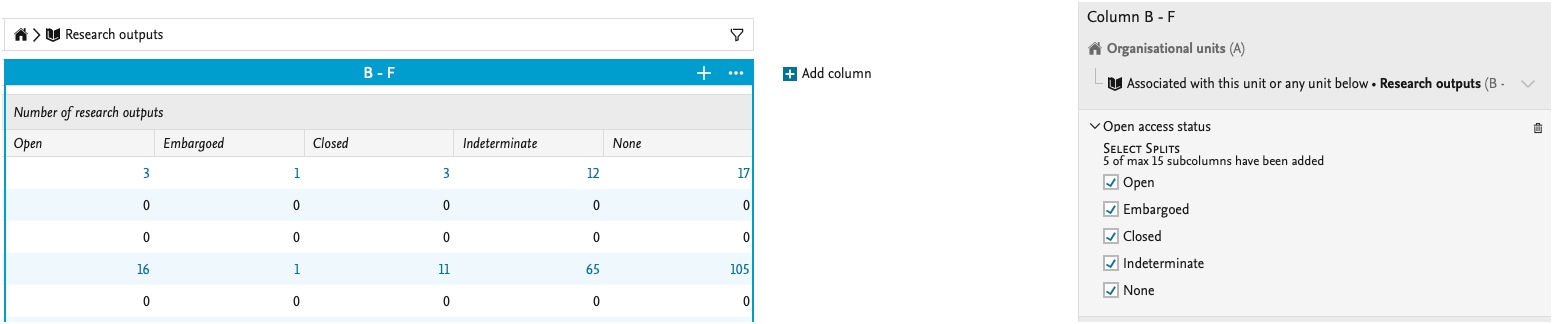
We have also added more values for the APC part of the Research output. You can now add information to the data table on the APC currency, whether the charge has been paid, and include the actual sum both in paid currency and Pure's default currency.
5.19
Open Access guidance section
We have introduced additional ways of communicating Open Access (OA) information on content. This includes details related to journals, publishers and/or an institution's own preferences. In the newly added 'Open Access Guidance' section of the Journal and Research Output editors, Administrators can now control what OA information is shown, presenting users with the most relevant OA references and guidelines at the right moment in their workflow. The section also includes Sherpa Romeo recommendations.
To configure visibility options for OA guidelines, Administrators can navigate to Administrator > Open Access > Open Access communication.
Institution's Open Access recommendations
Administrators can create an Open Access-specific message with hyperlinks to their institution's resources, and toggle the display setting to activate it on specific editors.
This message can be shown in the Journal editor, Research Output editor and the Electronic version upload editor.
Sherpa Romeo recommendations
Sherpa Romeo (SR) information has been updated (see the Sherpa Romeo section in these release notes for more information) and is now part of the 'Open Access guidance' section. Administrators can toggle the display setting to activate it on specific editors.
Sherpa Romeo information is always shown in the Journal editor, with control on visibility limited to the Research Output editor and the Electronic version upload editor.
Journal-specific additional information recommendations
Users with rights to edit journal metadata can add journal-specific OA information in a dedicated text field.
Journal-specific information is always shown in the Journal editor, with control on visibility limited to the Research Output editor and the Electronic version upload editor.
Configuration of Open Access guidance message and visibility on editors 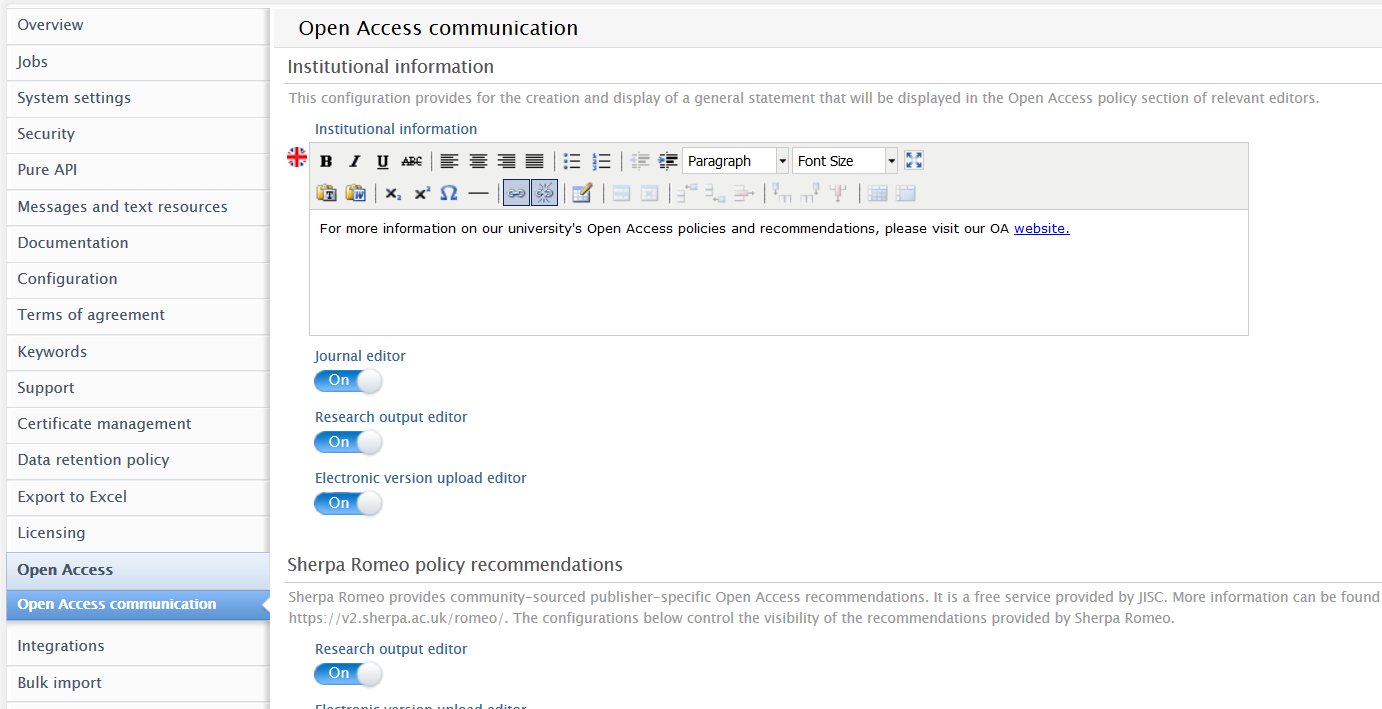
|
Open Access guidance in Journal editor Journal editor > Metadata 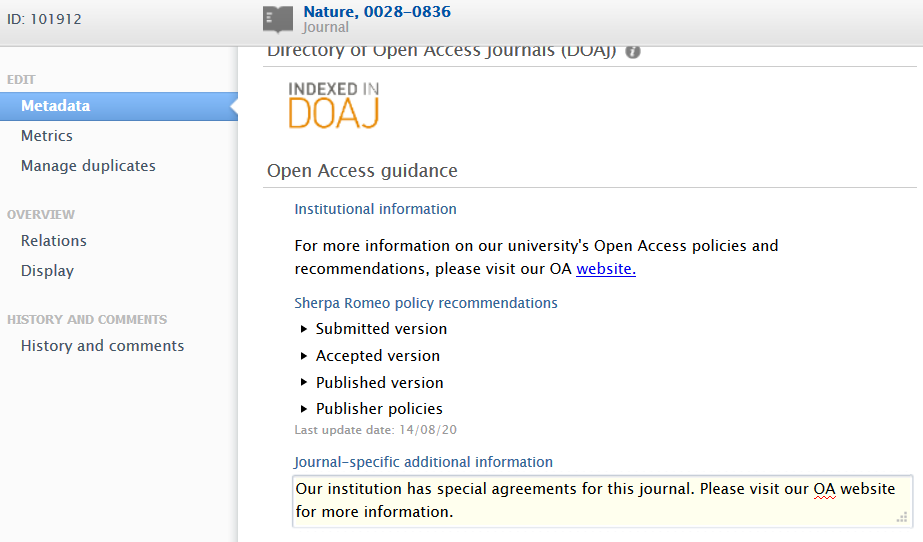
Open Access guidance in Research output editor Research Output editor > Metadata 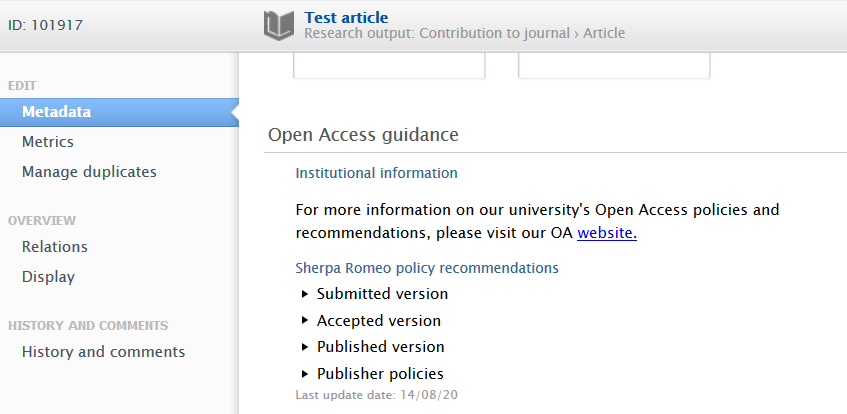
Open Access guidance in Electronic file upload window 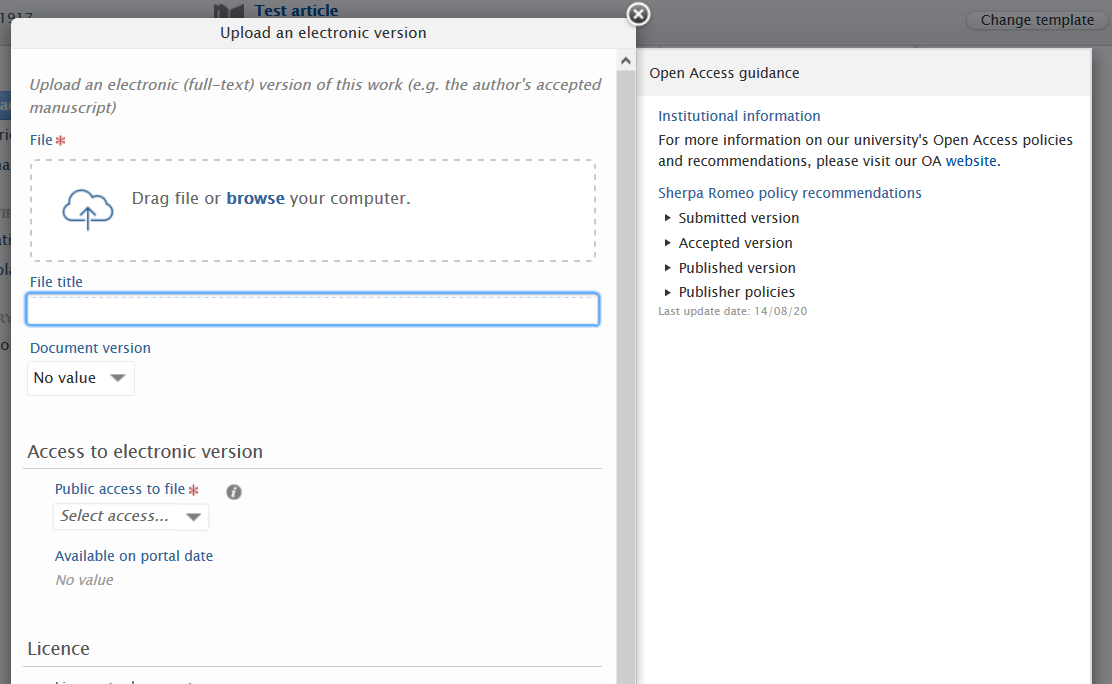
|
Open Access file upload reminder
To improve Open Access (OA) compliance, administrators can enable and configure reminder messages for users to add full-text files to content that is potentially OA. The reminder title and message body can also be customised to point users to their institution's own resources.
To enable the reminder
Administrators can navigate to Administrator > Open Access > Open Access communication and toggle on 'Set task and weekly digest email for personal users as default'.
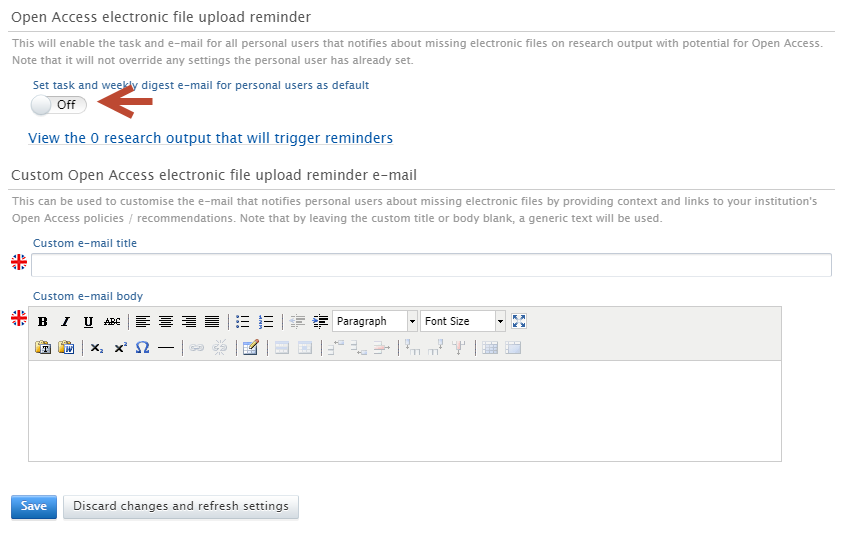
To configure the title and message body
The title and message body can be customised using the clearly labelled free text editors. Administrators can add URLs and rich text formatting to the message body if needed. If nothing is added to the title or description fields, an appropriate generic text will be added automatically.
Once the reminder has been enabled and configured, the configuration must be saved.
What triggers the reminder to be sent?
Reminders will be sent to the users who created the content when the following research output conditions are present:
The Open Access status is:
- None or Indeterminate
The Open Access policy is:
- Any Open Access route (based on Sherpa Romeo information)
And the Publication status filter is:
- Published, E-pub ahead of print, and Accepted/In-press
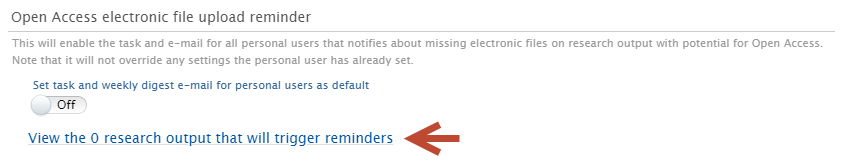
Administrators can examine the specific content that these conditions apply to by following the link 'View the X research outputs that will trigger reminders'.
Note: even if reminders are configured to be sent by Administrators, Personal users can still disable the reminders in their user preferences.
Sherpa Romeo model and API updates
Sherpa Romeo (SR) is a free service by Jisc (https://v2.sherpa.ac.uk/romeo/) that provides community-sourced publisher-specific Open Access recommendations. SR has recently transitioned from their color scheme to a new format which provides more information per publication status. Coinciding with the model changes are API changes including the need for customers to register for API keys.
Model updates
We have updated the display of SR information in Pure to reflect the model changes. Administrators can now control the visibility of SR information for the Research Output editor and Electronic version upload editor (see Open Access guidance section). The new format of SR recommendations provides more detailed information for each publication state of an article, i.e. Submitted, Accepted, Published.
You can check this detailed information on Research Output, Journal and electronic version upload editors, by clicking to expand each of the article state sections in Open Access guidance>Sherpa Romeo policy recommendations. The details include prerequisites, embargo periods, locations of the document, conditions, and more.
Note: Visibility on each of the editors must first be enabled.

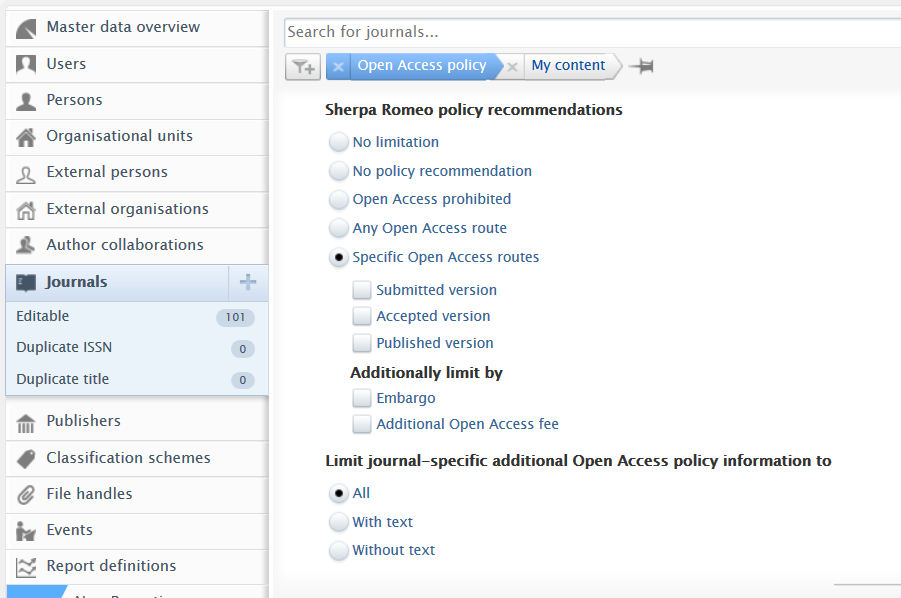
Sherpa Romeo filters
SR filters are now included in the Open Access policy filter set.
|
By default, the filter is set to No limitation, which means no specific SR filters are applied. Users can filter content by:
Note: The Limit journal-specific additional OA information filter will filter journals with or without user-provided additional OA information. This sub-filter does not directly affect SR data options.
|
API updates
A major difference, besides the model changes introduced in their API, is the requirement for an API key. Sherpa Romeo now requires customers to generate their own key, which can be done by registering with SR via https://v2.sherpa.ac.uk/cgi/register.
Once you have an API key you will need to configure the SR job in Pure. The job, Sherpa Romeo Journal Synchronisation, can be found in Administrator > Jobs > Cron job scheduling:
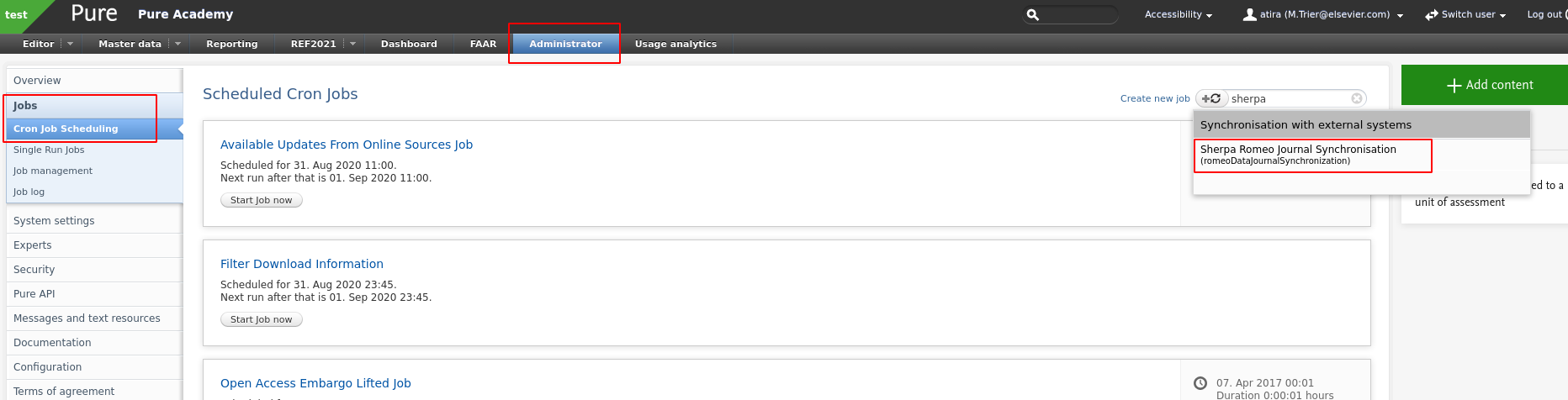
When you have found the job you will need to configure it accordingly.
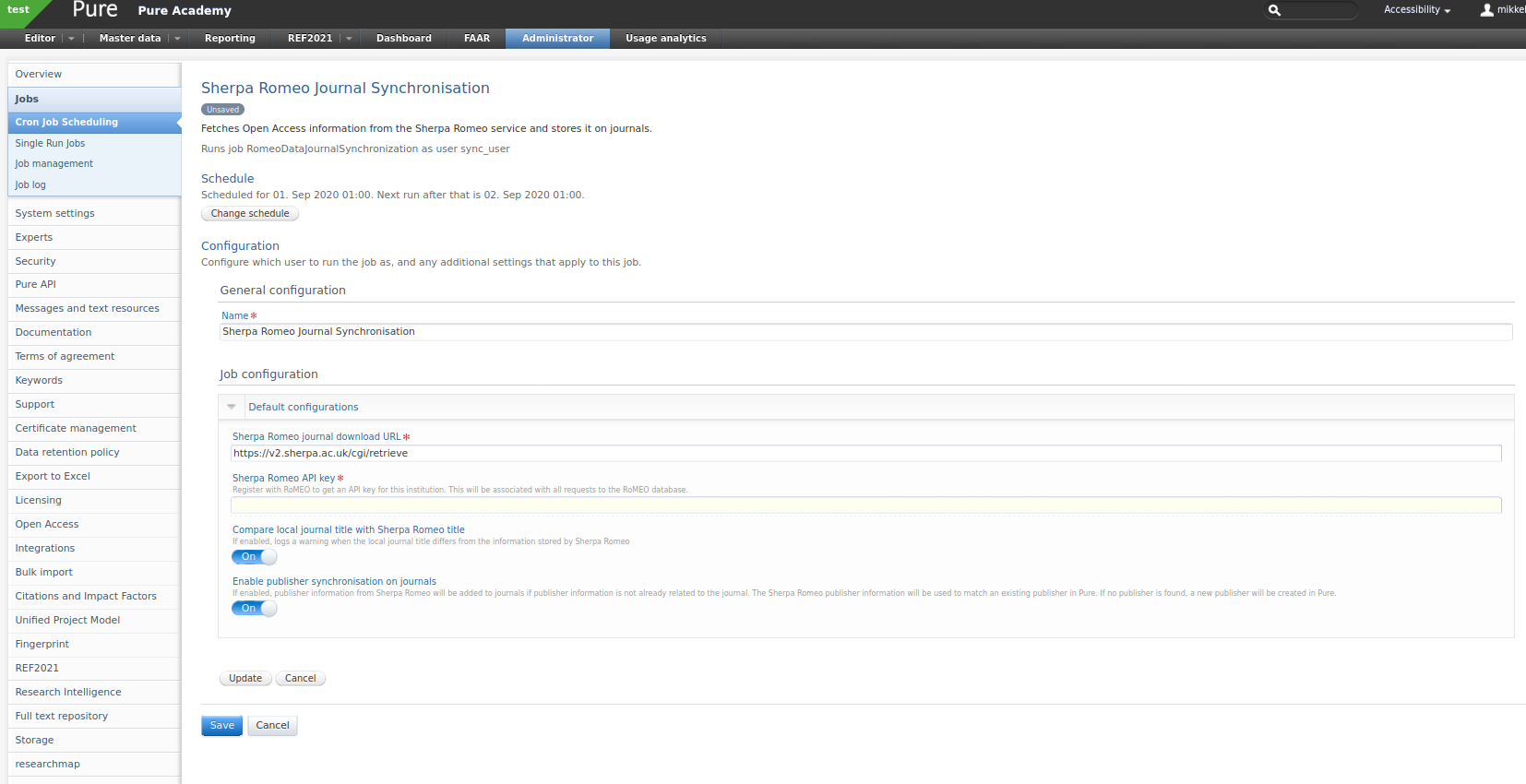
The following configuration values are available:
-
Sherpa Romeo journal download URL
- The URL used to retrieve data from Sherpa Romeo.
Sherpa Romeo API key
- Your institution's individual API key. You need to register for one (see above).
-
Compare local journal title with Sherpa Romeo title
- The job will match journals on ISSN. Enable this if you want the job to warn you when it detects a difference between the title in Pure and the one in Sherpa Romeo.
-
Enable publisher synchronisation on journals
- If enabled, publisher information from Sherpa Romeo will be added to journals if it is not already present. The Sherpa Romeo publisher information will be used to match an existing publisher in Pure. If no publisher is found, a new publisher will be created in Pure.
New Import Source: Unpaywall
Unpaywall (https://unpaywall.org/) is an Open Access database, harvesting content from more than 25,000 Gold OA journals, hybrid journals, institutional repositories, and disciplinary repositories.
The integration with Unpaywall supports researchers by expanding the range of available import data sources with an OA database indexing over 25 million publications, and providing researchers access to the best available online version of a record.
Instructions on how to enable, add and search for content from Unpaywall are shown below.
| Instructions | Screenshot |
|---|---|
| To enable Unpaywall as an import source, go to Administrator > Research Output > Import Sources | 
|
| To search and import content, go to ‘Add content’ and in Research Output > Import from online sources select ‘Unpaywall’. | 
|
|
Search for content using the search bar. It is possible to search by the DOI of the Research Output. It is then possible to import or remove import candidates. |
|
Instructions on how to add and configure Unpaywall as a source for Available Updates are shown below. For more general information on how to best configure rules for the Available updates feature, please read the Available updates section in the 5.13.0 release notes.
| Instructions | Screenshot |
|---|---|
| To enable automatic updates for Unpaywall, go to Administrator > Research Output > Import and Available Updates Configuration. | 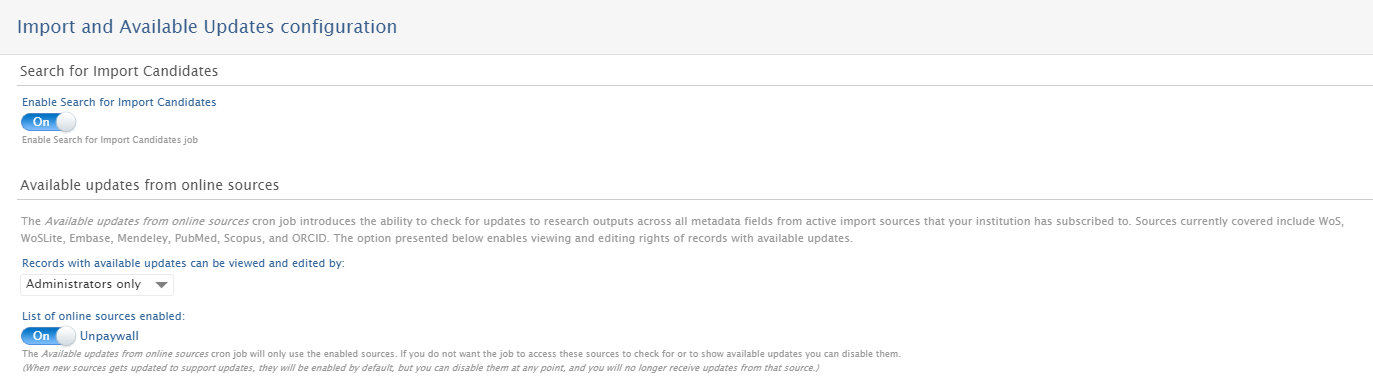
|
|
Update rules that determine how updates to specific metadata fields of a Research Output are handled, and Save configuration. Rules can be configured for Keyword Groups and URLs.
|
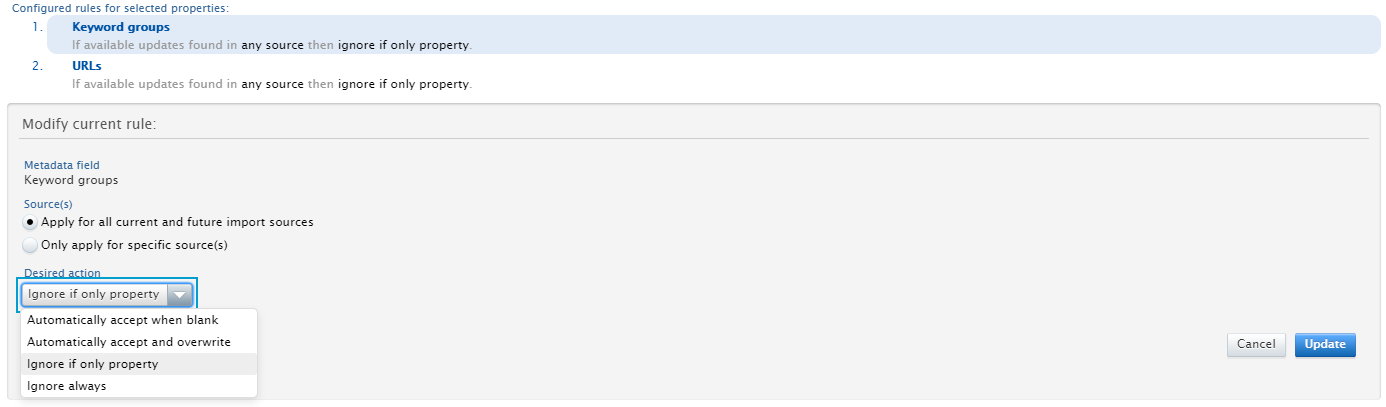
|
Note
By default, any available updates are highlighted in the Research Output Editor Window > Available Updates tab. You must manually select/merge the preferred options and save any changes.
New Import Source: CORE
CORE (https://core.ac.uk/) is an aggregator of OA repositories, harvesting research papers from data providers from all over the world, including institutional and subject repositories, Open Access and hybrid journal publishers. It contains over 186 million OA articles from tens of thousands of journals, collected from 9,980 data providers around the world. The full list of data providers can be found at CORE's provider listing. CORE will supply data for the UK REF 2021 Open Access Policy Audit to Research England, and the integration with CORE will make OA articles more easily accessible and more useful to researchers, and will allow institutions to monitor their OA output.
To enable CORE integration you will need to provide an API key, which can be requested at https://core.ac.uk/services/api/.
Instructions on how to enable, add and search for content from CORE are shown below.
| Instructions | Screenshot |
|---|---|
| To enable CORE as an import source, go to Administrator > Research Output > Import Sources | 
|
| To search and import content, go to ‘Add content’ and in Research Output > Import from online sources select ‘CORE’. | 
|
|
Search for content using the search bar. It is possible to search by author names, DOI and title. It is then possible to import or remove import candidates. |
|
New Import Source: bioRxiv
bioRxiv (https://www.biorxiv.org/about-biorxiv) is a free online archive and distribution service for unpublished preprints in the life sciences, containing over 80,000 papers by more than 348,000 authors.
The integration with bioRxiv provides researchers and institutions with content in biological sciences, especially relevant when following latest COVID-related studies and results.
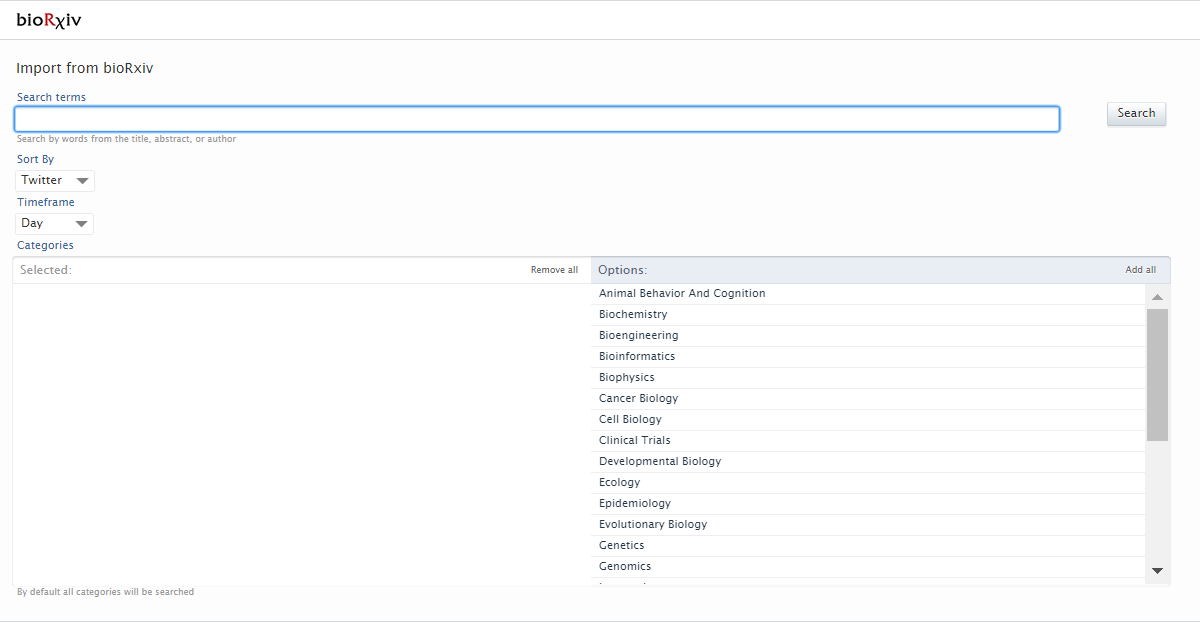
Instructions on how to enable, add and search for content from bioRxiv are shown below.
| Instructions | Screenshot |
|---|---|
| To enable bioRxiv as an import source, go to Administrator > Research Output > Import Sources | 
|
| To search and import content, go to ‘Add content’ and in Research Output > Import from online sources select ‘bioRxiv’. | 
|
|
Search for content using the search bar. It is possible to search by author names, DOI and title. You can define the time frame and subject category in which you want to perform the search. It is then possible to import or remove import candidates. |
|
OA information for Scopus and CrossRef content
In this release, we have enriched the data of Research Outputs imported from Scopus and CrossRef by retrieving additional Open Access information.
Records imported from Scopus will now contain additional information on:
- Open Access status flag
Records imported from CrossRef will now contain additional information on:
- Open Access status flag
- License
Information on the Open Access status of a document is associated to the DOI, electronic version, or to a specific version (file) of the document.
5.18
Jisc Publications Router Integration
In this release we have added integration to the Jisc Publication Router. The Jisc Publications Router provides a way for Pure customers to receive notifications from publishers whenever a publication with their affiliation has been added or updated. By bypassing indexing services such as Scopus or WoS, the Jisc router speeds up the import of relevant content into Pure.
In addition to reducing your Institution's administrative effort and maximising discovery and distribution of content, The Jisc Publications Router helps institutions comply cost-effectively with the Open Access policies of research funding bodies, currently valuable to UK customers considering the upcoming REF.
Access to the Jisc Publications Router is currently limited to UK customers. Before using the router integration in Pure, your institution will need to have an account set up and an agreement in place with Jisc – details available at https://pubrouter.jisc.ac.uk/about/start-up-guide-for-institutions/
Setup instructions
| Instructions | Screesnhot |
|---|---|
| To enable the integration with Jisc Publications Router, in Administrator > Research Output > Import Sources set the 'Jisc Publications Router integration' to ON. | 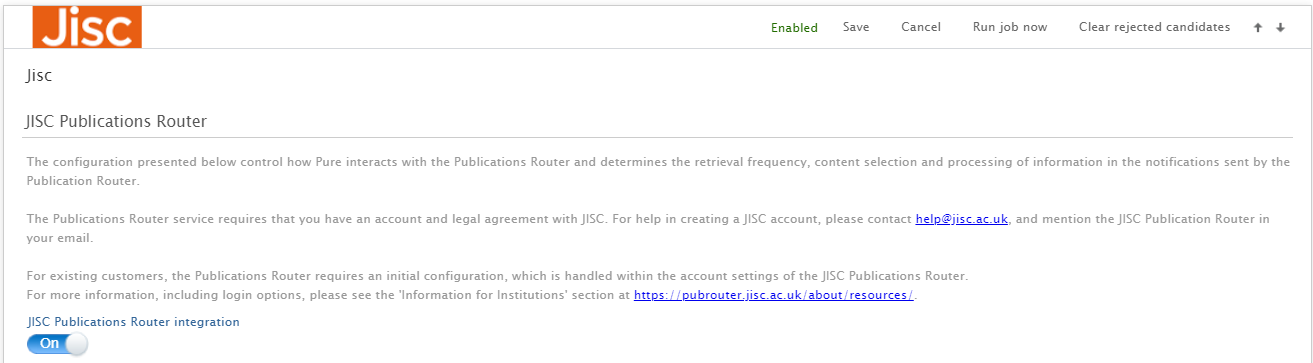
|
| In the 'Configuration' tab insert Account ID and API Key: | 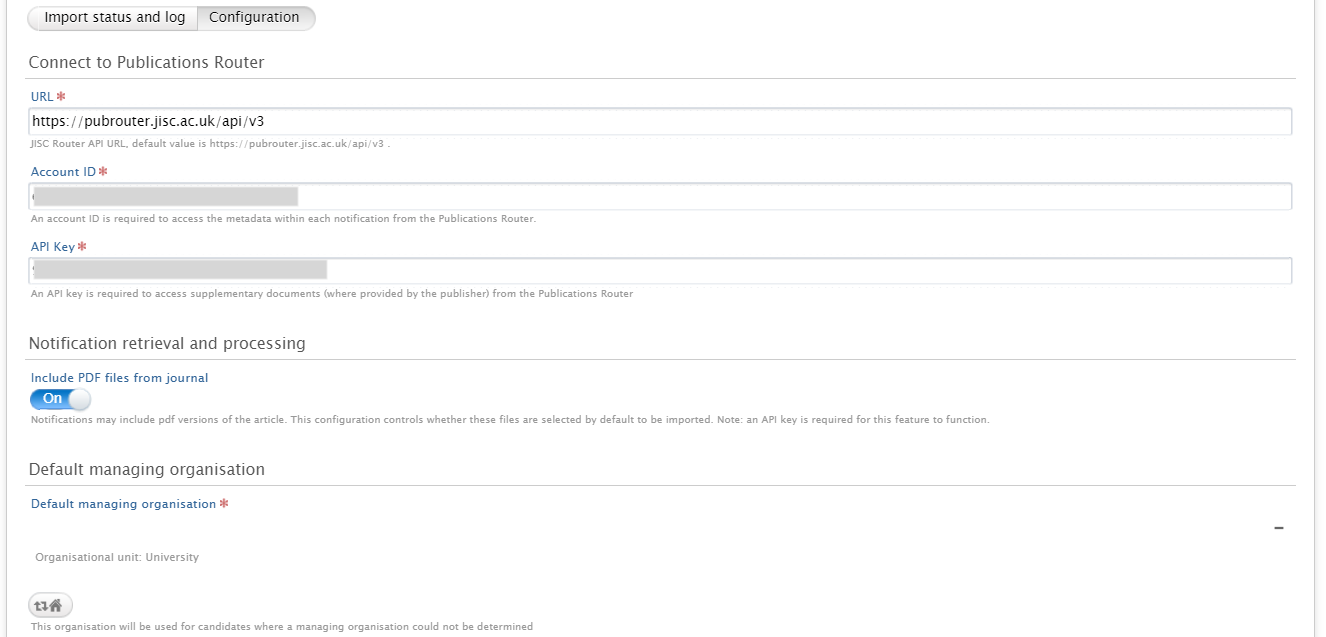
|
|
By default 'Notification retrieval and processing' is turned ON, meaning that if a pdf version of a record is found this will automatically be imported. It is possible to toggle this option OFF; in this case when a new import canddate is found it will still be possible to select the attached pdf file, when available. Once the integration has been setup, the job will run in the background and find import candidates. This job is configured to run once per day, between 06:00 and 08:00 (the exact time is randomized as to stop all Pure instances to begin querying the Jisc API at the same time). The job can also be triggered manually by selecting 'Run Job now' |
|
Import and Update Candidates
Once the integration has been enabled, import candidates can be found in Research Output > Organizations with import candidates. Records will be shown for the default managing organization.
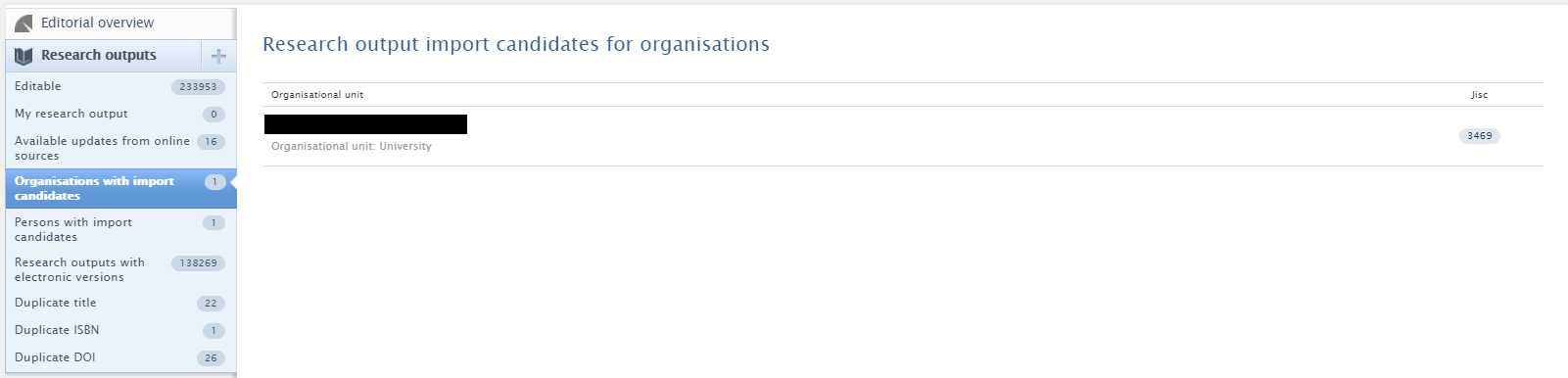
Update candidates are created if a match is made on either DOI or PubMed ID. It is then possible to import or reject candidates. Once candidates have been rejected, they will not be shown when the synchronization runs again.
Rejected candidates can be cleared by clicking "Clear rejected candidates" on the Jisc Router job configuration.
Sherpa Romeo API updates
Sherpa Romeo is deprecating their current 'color' service for journal OA policies and permissions. Based on current information found at http://sherpa.ac.uk/romeo/index.php, and confirmed by Jisc, the providers of Sherpa Romeo data, the following dates are confirmed:
- Launch of new service: April 14th 2020
- Decommission of current service: July 31st 2020
The updated web access can be found here: https://v2.sherpa.ac.uk/romeo/
Significant changes have been made to Sherpa Romeo, with multiple options for Open Access routes per publication state. Interviews with customers are underway to determine what is the best information to surface at the research output and the journal content level.
Service to Sherpa Romeo will continue to be available until the API shutdown, upon when Pure will switch off access to the old API to prevent system errors.
In 5.18.1 users will be shown a link to Sherpa Romeo to access the latest journal OA routes, and once customer interviews are complete, the updated information will be surfaced in full on the research output and journal editors. The transition to the new Sherpa Romeo will be complete on release of 5.19.
We highly recommend administrators and other relevant roles review the new OA access routes information.
5.16
Use of journal or approximated dates for content from Scopus
Scopus provides publication data with all content indexed in Scopus. Occasionally, dates provided by the journals, and shown on Scopus.com, are vague - for example 'Spring 2019', or 'January 2019' or '2019'. Scopus provides an approximation service that fills in 'missing' month and day information. As a result, content with a journal-provided date such as March 2019 appears in Pure as '1st of March 2019'.
To ensure that the content found in Pure best reflects the date found in the journal, we have introduced a configurable feature. This allows administrative users to:
- configure content imported from Scopus to use the exact journal date versus the approximated date,
- backfill content already imported from Scopus.
The feature will not overwrite any manually changed publication dates, and is disabled by default.
Warning for REF customers
Before enabling this feature and/or back filling content, REF customers should note: Running the run once job could change the publication date on content that has been proposed to REF2021, in some cases removing the full publication date. This might have implications in relation to REF compliance. We recommend to first run the job on your test servers to see the implications of the job in relation to REF2021.
Example record from Scopus.com

Provided below is an example of the publication date as seen in Scopus.com for the record with Scopus ID: 85058052904. The publication date, as of the writing of this release note text, is January 2019 and reflects the journal's representation of the publication date. However, the publication date when imported into Pure is represented as '1 January 2019' due to the data provided via the Scopus API.
| Configuring Scopus as an import source for future content import: | |
|---|---|
|
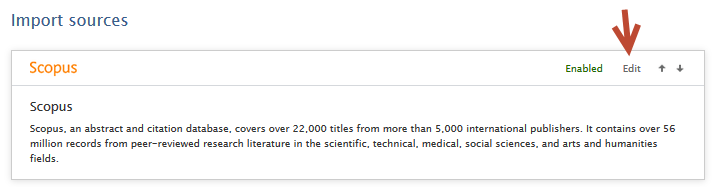
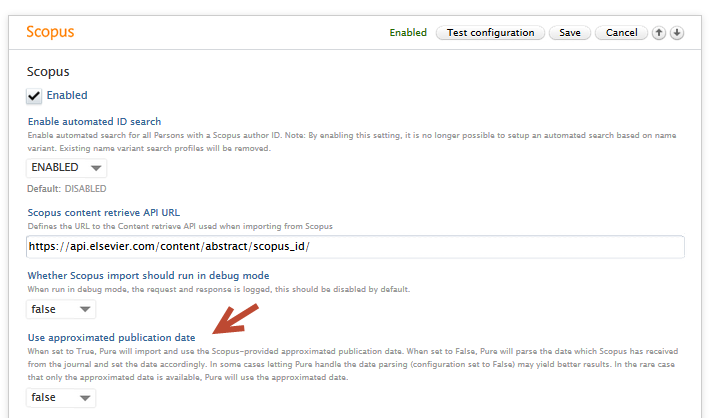
Note: For imports from Scopus, you must have enabled Scopus as an import source.
|
1. 
2. 
|
| To use exact journal dates on all previously imported content: | |
A single run job has been created that will convert all content previously imported from Scopus to use exact journal dates. This job will not overwrite any manual publication date changes even if the content was imported from Scopus. To use the job:
If you convert all previously-imported content to use exact journal dates, you will not be able to use the same job to convert back to using approximated dates.
|
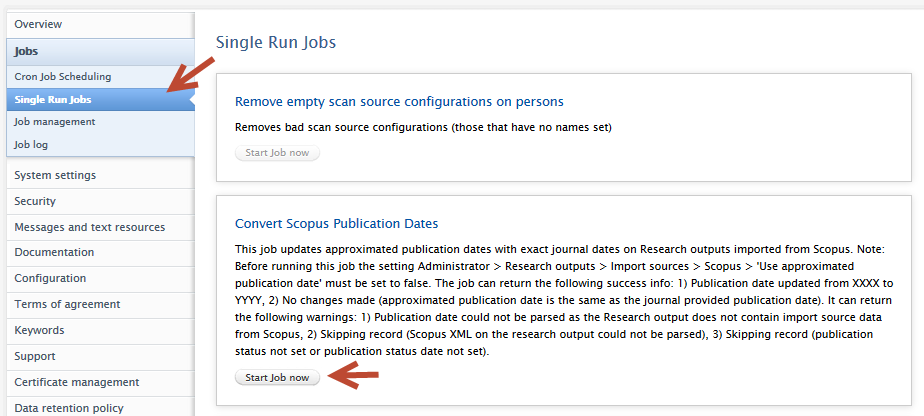
|
|
Once the job has run, you can inspect the results in the job log by clicking on the job run date. Users can use the dropdown filter and search option to view successes, or warnings if records were not processed. Success status information includes:
Warning status information includes:
|
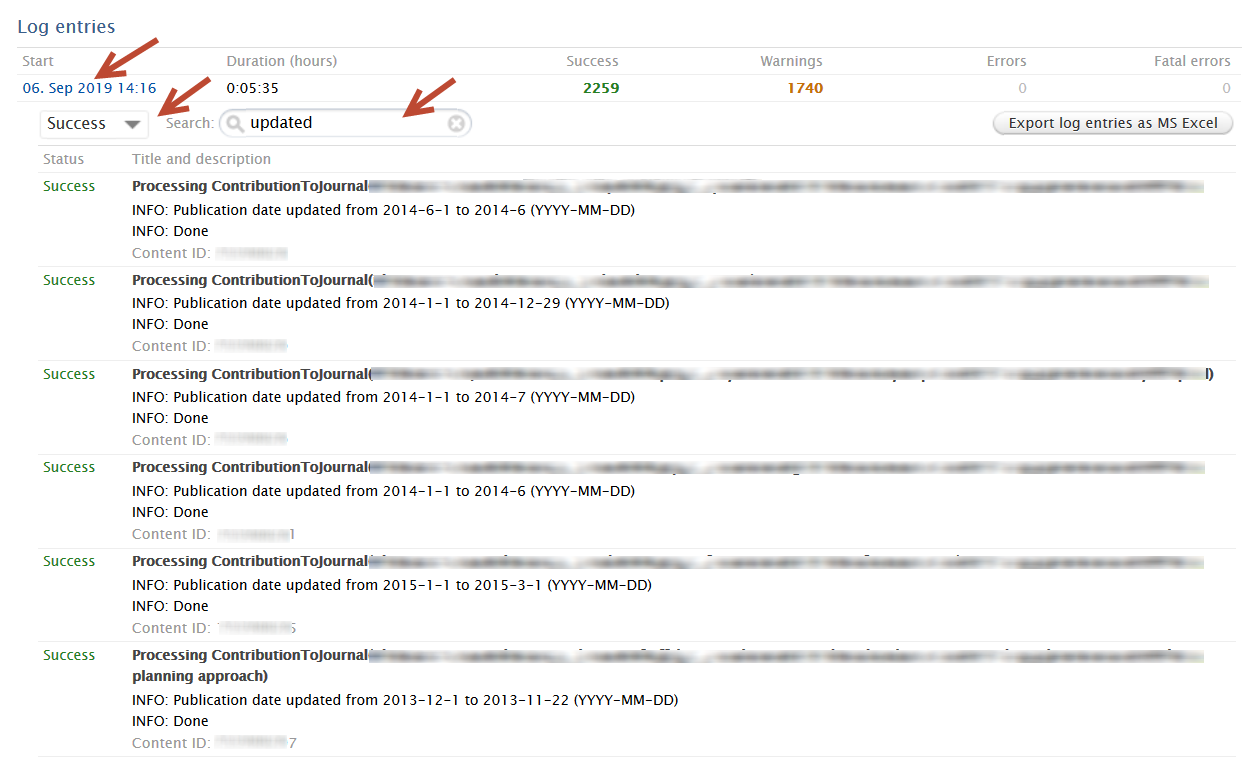
|
| If you are a QABO customer: | |
|
To ensure all content imported from Scopus uses approximated dates:
To ensure all content imported from Scopus uses exact journal dates:
|
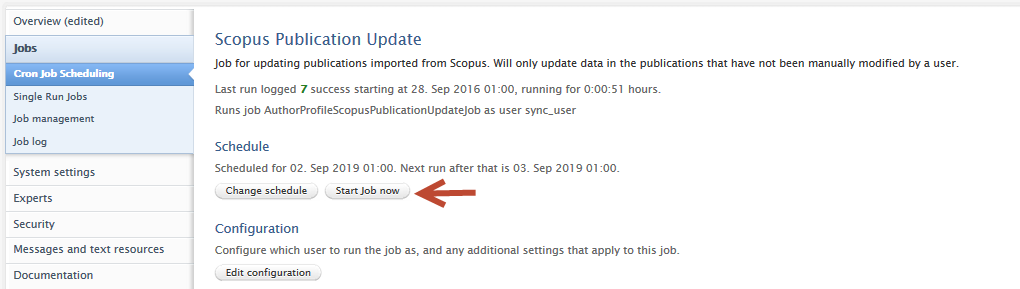
|
|
Important note for QABO customers: if you would like to process all previously-imported Scopus content, we suggest you enable 'Force update of all publications from Scopus' in the job configuration. Please remember to save any changes to the job configuration. |
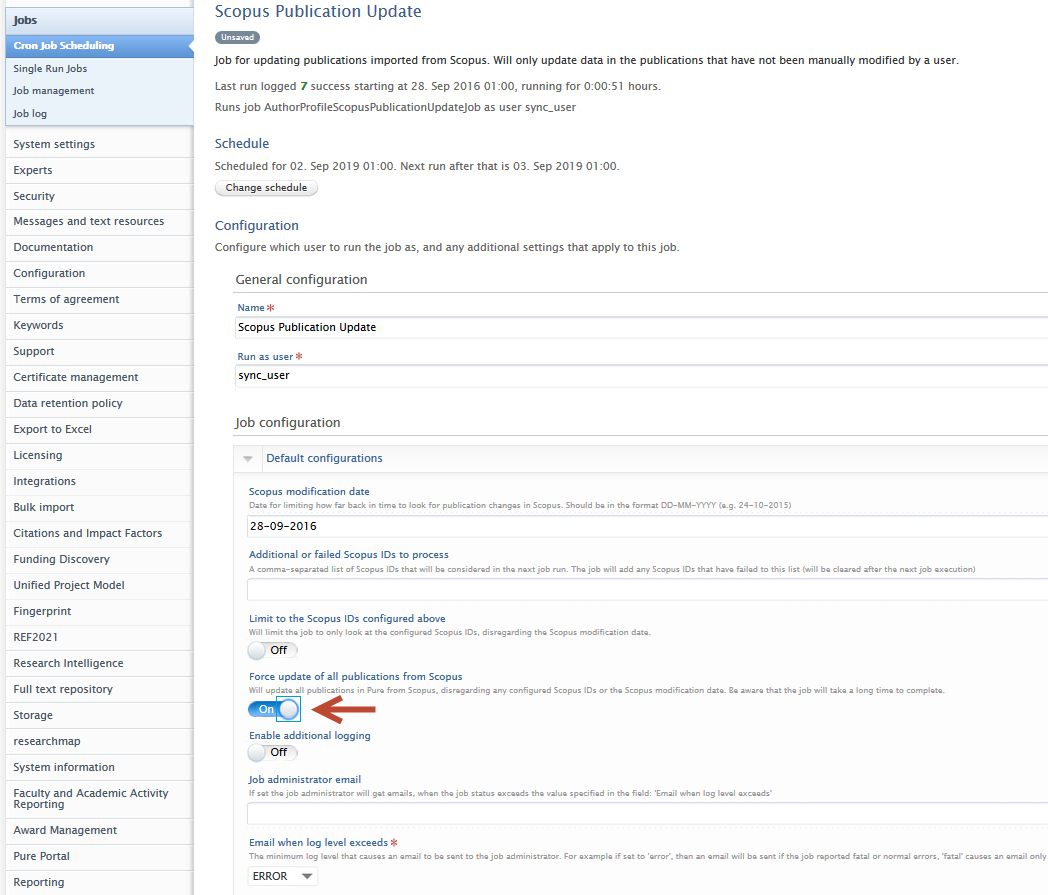
|
5.15
Showing metadata on embargoed documents - Portal
Previously, if a document attached to a research output was under an active embargo, all information pertaining to the embargoed document was hidden.
This doesn't make much sense as, even though the document of course should not be made available, it can be very helpful to the user to know:
1) that there is an embargoed document attached
2) the duration of the embargo and
3) the title of the document,
This information could potentially even help the user retrieve the document from another source, for instance via a "Request for copy" to the author.
These three fields will now be shown for embargoed documents on the Pure Portal. This provides more utility to the Portal visitor, while still obeying the terms of the embargo.
Please note: This change does not affect "Restricted" documents.
5.14
Logic and behavior rules for Available Updates from Online Sources
Property logic and behavior rules for the 'Available updates from online sources' job have been updated and refined, to enable users to be more selective of data received from different online import sources and to changes made to metadata fields.
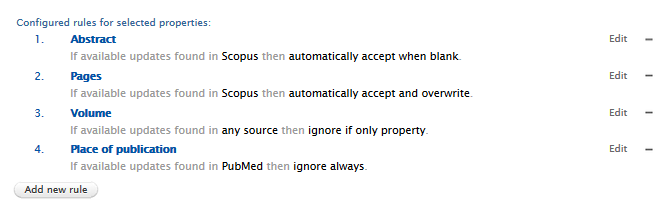
Administrator users can now define rules that determine how Pure automatically handles updates to the metadata fields of a research output.
If the 'Available updates from online sources' job is enabled, and no rules are set, by default any available updates are highlighted in the Research output editor window > Available updates tab, and the user must manually select/merge the preferred option(s) and save any changes.
Please note:
General logic: Rules follow an 'if-this-then-that' logic - a user selects a metadata field, an action to perform on this field, and the import source(s) the action must take into account.
Where in configuration: Rules can be set in Administrator > Research outputs > Import and Available Updates configuration > Property logic and behavior rules for 'Available updates from online source' section.
Import sources: The import sources that are available for the rules to function are determined by what is enabled in the 'Available updates from online sources' section (available via the same path as above).
Instructions for use:
| Adding a new rule: | |
|---|---|
|
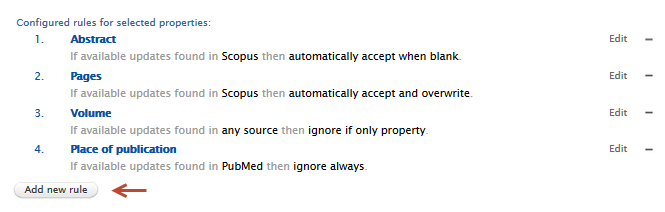
|
| 2. Select a field to be acted on in the 'Metadata field' drop down menu. | 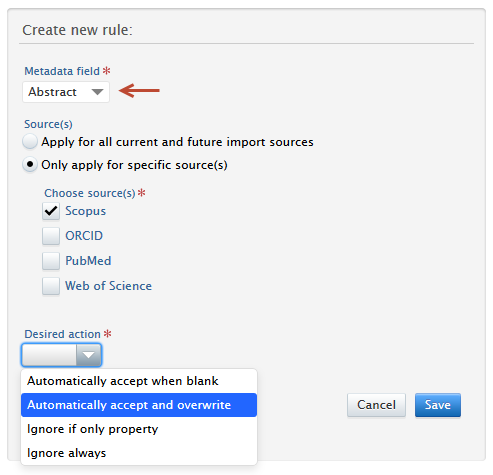
|
|
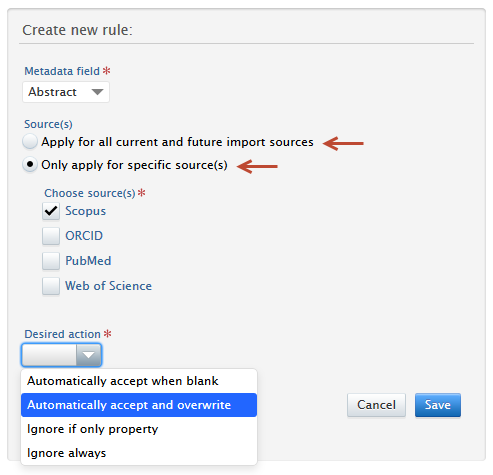
3. Choose either:
|
|
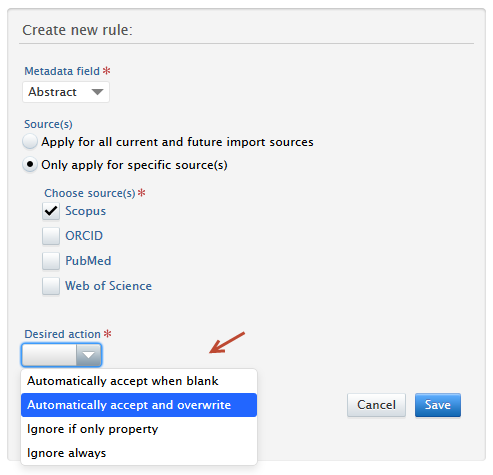
4. Select an action:

|
|
| Editing or removing a current rule | |
|---|---|
| A rule can be edited by clicking on the 'Edit' button next to each rule. | 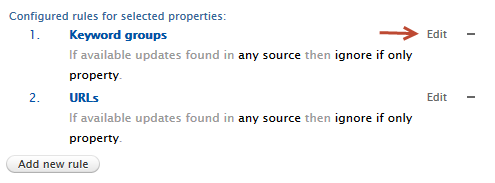
When a user is modifying a current rule, the rule in question is highlighted in the list, and only the Source(s) and Desired action options are allowed to be changed. 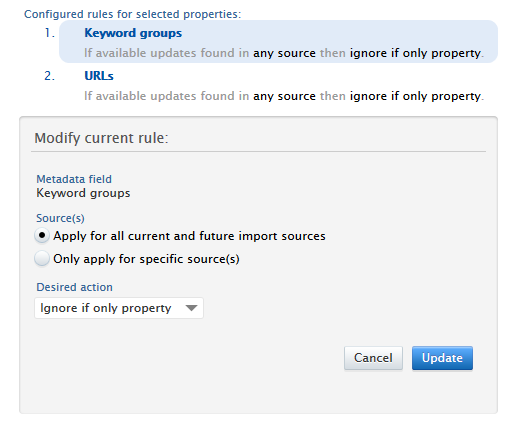
|
| A rule can be removed by clicking on the '-' button next to each rule. | 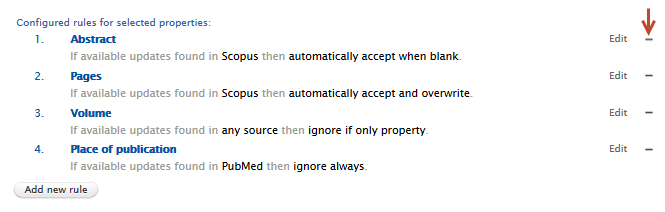
|
Filtering research outputs affected by Available Updates from Online Sources job
| Filters | Screenshots |
|---|---|
|
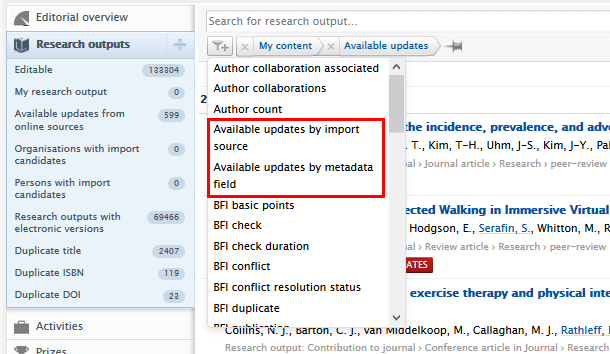
Users can filter research outputs that have available updates by import source, and by metadata field. Using these filters, users can pre-select and inspect records before processing them either manually or via rules. Users can select one or more source or metadata field.
Important note: After enabling the job, it may take up to a week for all data to be scanned and potential updates identified. |
|
Note
Care should be taken when using these rules to avoid creating conflicts between multiple actions and sources. If a new rule conflicts with other rule(s), one or more of the following conflict messages will be displayed, and the suggested courses of action are explained below.
| Conflict error message | Suggested course of action |
|---|---|
| Required fields are missing. | Fill all fields to create a rule. |
| Rule is configured for both all and specific sources. | Only select 'All current and future sources' or specific source(s) |
| Automatically accept actions can only be set for one specific source. | Use Automatically accept actions for only one source in case there are different data between sources |
| An action is already configured for '{field}' for this source: {source}. | Check current rules and modify if necessary so that only one action per metadata field and source combination exists. |
| An action is already configured for '{field}' for the sources: {source}. | Check current rules and modify if necessary so that only one action per metadata field and source combination exists. |
| A rule is already configured for '{field}' using an Automatically accept action. A metadata field can only be configured to Automatically accept for one source. | Check current rules and modify if necessary so that only one source is used for an Automatically accept action. |
| A rule is already configured for '{field}' using the same action but a different range of sources. A metadata field can only be configured to use either all sources or specific source(s). | Check current rules and modify if necessary so that only one action per metadata field and source combination exists. |
Import sources and rule
If a source is disabled in the section 'Available updates from online sources', an warning message will is shown indicating which rules will be affected by disabling the source. A user may save this configuration if they wish but the source will be shown as disabled in any rules affected.
| Disabling sources | Screenshot |
|---|---|
| When a source is disabled and this affects a rule, a warning message is shown, specifying which rules it affects. | All sources enabled: 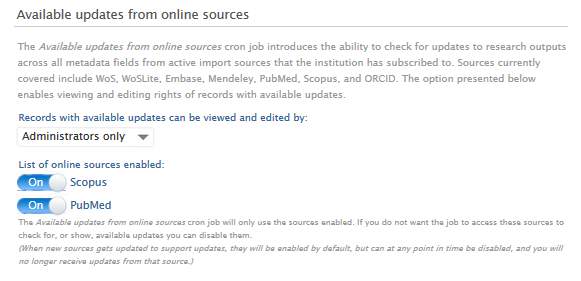
Source disabled, with effect on rule(s): 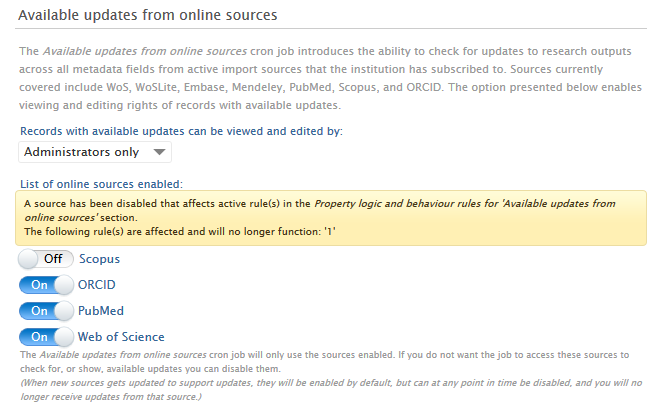
|
|
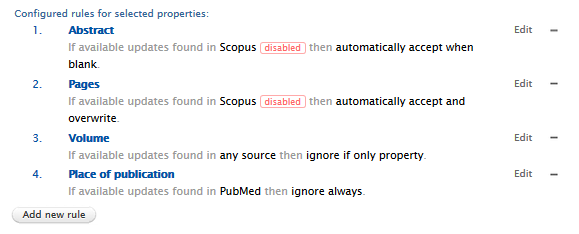
If the settings are saved, a 'disabled' tag will appear in the rule(s) next to the source(s) affected. The rule will no longer function as intended as the source will be excluded from the logic of the rule. Note: If a source has been disabled but a rule uses the 'any source' option, no error or 'disabled' message will be shown. |
|
Once rules are added or modified and the configuration page has been saved, the rules (and actions specified in these rules) are set to work on the current data. To avoid conflicts between current rules and rules being modified and added, a user cannot add or modify rules until the current set of rules have completed their processing of the current data. A warning message will be shown (see below).
Once the processing has been completed, rules may once again be added or modified.
5.13
Research outputs: Available updates from online sources
As part of our continuing focus on increasing automation to reduce the burden on our users with regard to data entry into Pure, this release introduces new functionality to update existing Pure Research Output records with new and potentially more complete metadata from available online sources. The feature searches online sources for updated information on existing Research Outputs in Pure and presents users with the updated data for consideration.
By default, this feature is ENABLED and updates are by default only presented to Administrators - except for PRS customers using the QABO 'Scopus Publication Update Job', where the feature is by default DISABLED. Further information for PRS customers is available below.
While updates from online sources are pending consideration, Research outputs can still be edited and saved in the normal way, but with some restrictions:
- For users not enabled to access the available updates from online sources (e.g. Personal users), all metadata fields in the Research output editor will continue to be editable while updates from online sources are pending consideration
- For users enabled to access available updates from online sources (e.g. Administrator), those metadata fields with updates pending consideration will not be editable in the editor. Should the user wish to modify fields with updates pending consideration, the user will first need to address all fields with available updates
Presentation of available updates from online sources
When updated metadata is found for existing Research Output records in Pure (as per the configuration and matching details provided below), Administrators are presented with a list of Outputs with available updates. For each Output, the Administrator can then review these updates and decide the appropriate action (accept, ignore, merge).
The list of Research outputs with available updates from online sources is accessible through the menu under Research outputs. 
|
Alternatively, records with available updates can be filtered using the filter options in the Research outputs window. Also available is the ability to filter available updates by import source. 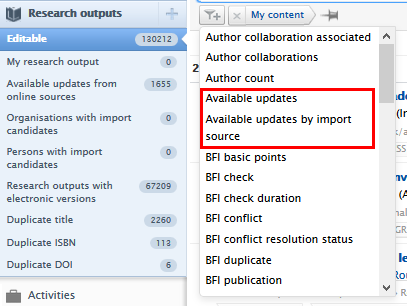
|
Note
If the only updates available are for the Keywords and Other links fields, the updates will not be presented to the user for consideration and the fields will not be updated. If updates are found for any other fields, the available updates will be presented to the user for consideration.
Acting upon available updates
To enable authorized users to consider the available updates from online sources, a new 'Available updates from online sources' window of the Research output editor has been introduced, within which the user can choose to accept, ignore, or merge each available update.
Upon selecting an Output with available updates from online sources, the Research output editor opens to a new 'Available updates from online sources' window of the Research output editor.
Differences in metadata fields between the existing Pure Research output record and the import source(s) fields are highlighted in yellow.
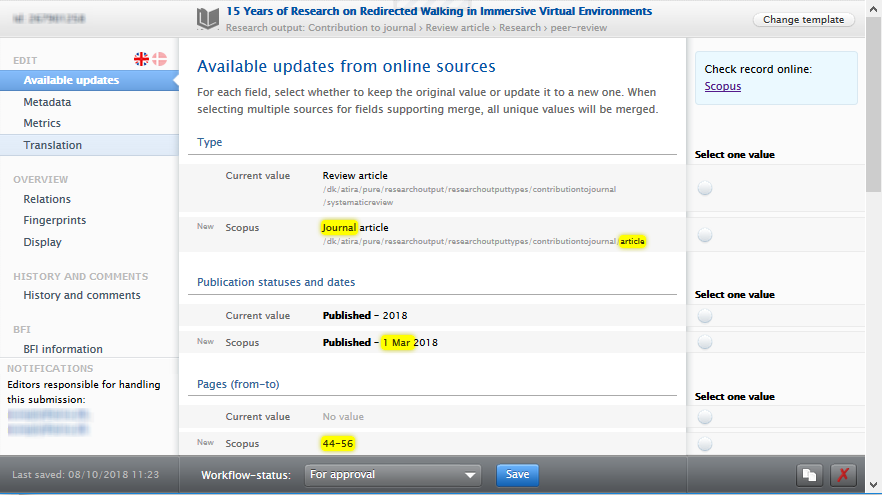
- For fields in which the Pure Research output record is null or blank, the value from the import source(s) is pre-selected.
- Users can select from each of the options presented for each metadata field.
- Once selected, the chosen value is highlighted and non-selected values are crossed-out.
- For some metadata fields, the option to merge values is available.
- Updates relating to the Journal field are not available for selection, even if there are differences between the Pure record and the import source(s). This is to ensure that such a fundamental change can only be implemented by directly editing the affiliated Journal in the Research output record itself, and to ensure that master Journal records are not unexpectedly altered.
Note: To edit metadata fields that do not have available updates, simply click the Metadata menu tab and all normal editing options are available.
Viewing current publisher or database version of record
If the user would like to view the most current version of the record, as presented by the publisher (via the DOI) or database, a link is provided at the top of the window.
Viewing current publisher or database version of record
If the user would like to view the most current version of the record, as presented by the publisher (via the DOI) or database, a link is provided at the top of the window.
Search, matching, and comparison processes
The search for available updates is based on unique identifiers in the Pure Research Output record. These are:
- Scopus publication IDs
- DOIs
- ISBNs
- ORCID ID*
* For those Research outputs with an ORCID ID (i.e. imported from ORCID - see below), the ORCID ID will be used to check ORCID for any updated data.
Provided the Pure Research output is within the parameters determined in the configuration settings (see below), the job uses these IDs to check enabled import sources. Once the IDs have been matched to records in each import source, the metadata fields in the Pure Research output are compared against the available metadata fields in each import source.
- For fields in the Pure Research output that are null or blank, all available non-null data for those fields are retrieved.
- For fields in the Pure Research output record that have data, these fields are compared using a comprehensive string or number matching process. Wholly different values and minor differences in spelling or extra or missing characters trigger the fields to be presented to the user in the update candidates user interface.
Publication statuses and dates
For publication dates, care has been taken to only suggest updates that are more refined or wholly different. For example:
| Source A | Source B | Source C | Source D |
|---|---|---|---|
| Current published date | |||
| 2 June 2018 | |||
| Data held by source | 2018 | 2017 | |
| Outcome in Pure | Not suggested as an available update | Suggested as it is wholly different | Not suggested as it is less refined than the existing value |
The publication status of records is also considered when checking for updates. For example, where a record has a current publication status of 'ePub ahead of print', any other statuses (and dates) found from online import sources will be presented to the user. If a user selects an additional status and date, this status and date will be added to the record, alongside the existing status(es) and date(s). Existing publication states will not be overwritten.
Electronic versions (files, DOIs, and links)
This feature will not modify or remove existing files or links held on the record. Existing DOIs will be checked against the available online sources and modifications will be suggested in the same way as other metadata updates are presented. Where a DOI update is selected, default values for related fields are as follows:
- Document version = Final published version
- Public access to file = Unknown
- License to document = Not set
Supported sources
The 'Available updates from online sources' feature supports the receipt of updates from the following sources:
- Web of Science
- Web of Science (lite)
- Embase
- Mendeley
- PubMed
- Scopus
- ORCID
Only those sources that are enabled as import sources will be included in the update search.
The list of sources will increase over time and notifications of new additions will be included in the relevant release notes.
There is currently no means of ordering or prioritising updates from the different sources; improvements to this are expected in upcoming releases.
Configuring the Available updates from online sources feature
The 'Available updates from online sources' feature is delivered in Pure as a cron job, with configuration options such as:
- scheduling
- years to search
- authorised user roles
- exclusion of individual import sources.
The 'Available updates from online sources' feature can also be removed from the schedule.
The 'Available updates from online sources' job can be added to the schedule by selecting it in the drop down list Administrator > Jobs > Cron Job scheduling > Select job from Create new job dropdown list. The job can be found under the 'Synchronisation with external systems' section.
By default, this feature is ENABLED, except for PRS customers using the QABO 'Scopus Publication Update Job', where the feature is by default DISABLED. Further information for PRS customers is available below.
Configuration options
The 'Available updates from online sources' job can be configured as follows:
Starting year
The job is configured to only consider Research outputs published in the specified year and later. The default value is the current year. For example, configuring the Starting year as 2017 will result in only Research outputs published in 2017 being considered.
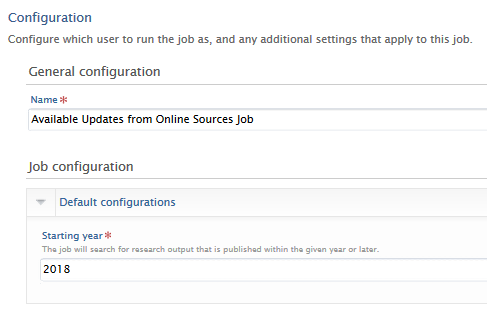
This configuration can be accessed via Administrator > Jobs > Cron Job scheduling > Available Updates from Online Sources Job > Edit configuration > Default configurations.
Access to results
This feature is, by default, only available to Administrators. But additional user roles (Editors and Validators) can be given access via Administrator > Research outputs > Import configurations > Available Updates from Online Sources.
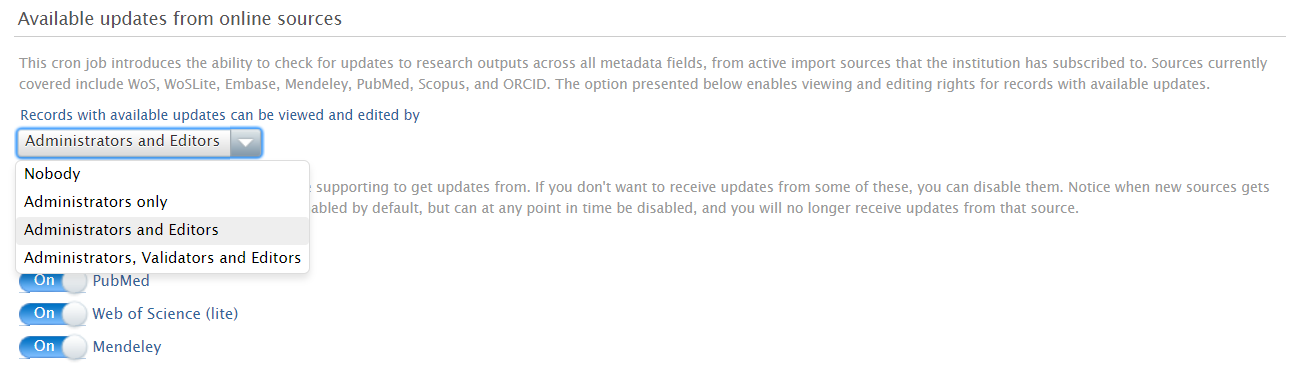
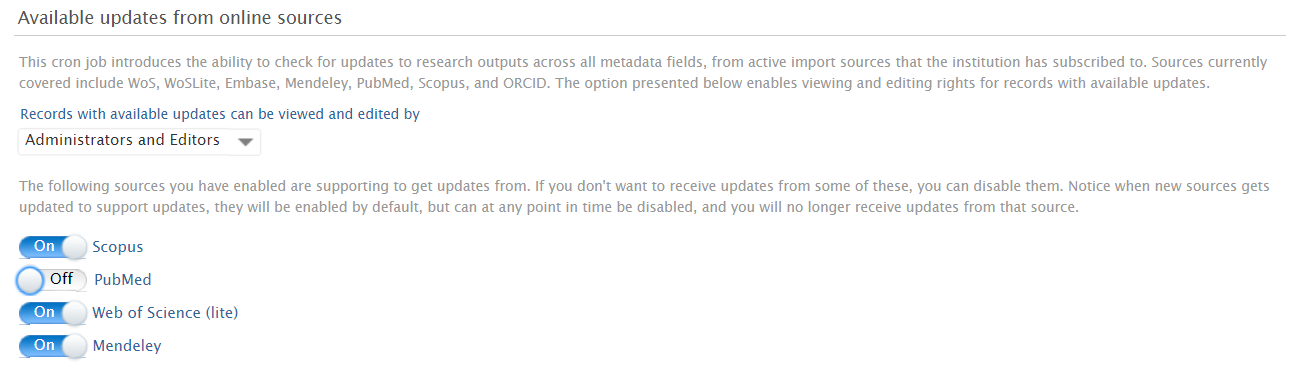
Enabling / Disabling individual online sources
Online sources can be excluded from the feature by toggling on or off for the individual source at Administrator > Research outputs > Import configurations > Available Updates from Online Sources (by default, all enabled sources are included). Note that the list of online sources available is limited to those enabled as import sources (as per Administrator > Research outputs > Import sources).
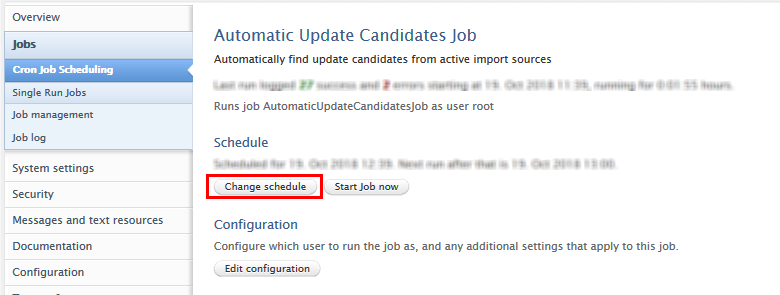
Removing 'Available updates from online sources' job from schedule
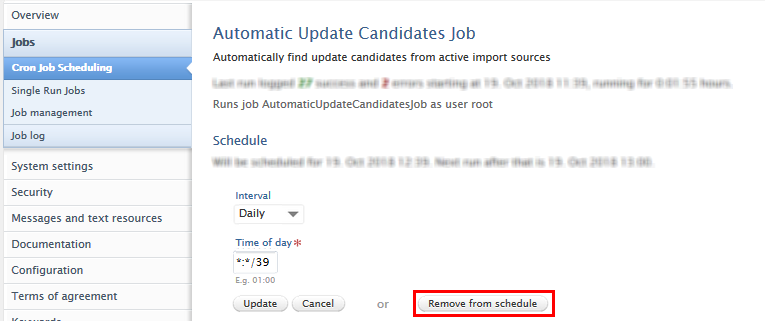
The job can be effectively disabled by removing it from the schedule. This is done by opening the job via Administrator > Jobs > Cron Job scheduling > Available Updates from Online Sources Job
Once the job is open, Change Schedule > Remove from schedule.
PRS customers
Records updated by normal use of the Available updates from online sources feature will affect the functioning of jobs specific to PRS customers that rely on Scopus as an import source.
Due to differences in the default scheduling of, and the data state against which checks are made, the Available updates for online import sources (daily/live data) and the QABO Scopus Publication Update Job (weekly/snapshot data), and the aggressive matching processes of the Available updates for online import sources job, it is highly likely that the suggested updates from Available updates for online import sources may precede updates added by the QABO job. If any of the suggested metadata updates are accepted through normal use of the Available updates for online import sources job, the QABO job, as designed, will not look for updates on the specific metadata fields of the records that were updated or modified by the Available updates for online import sources job.
Recommendations:
- For PRS customers that rely on the QABO Scopus Publication Update Job to update records from Scopus, it is recommended not to enable the Available updates for online import sources job to avoid any inadvertent modifications.
- If PRS customers would still like to enable the job, it is recommended that Scopus is excluded from the list of import sources to be used by the Available updates for online import sources job (see section on Configuration options).
- If PRS customers would still like to enable the job, and have Scopus as an import source to be used by the Available updates for online import sources job, please contact the Pure Support team.
5.11
SSRN: New Research Output import source
We are delighted to announce that we have added the Social Science Research Network (SSRN) as an online import source for Research Outputs.
SSRN is a worldwide collaboration of over 352,400 authors and more than 2.2 million users that is devoted to the rapid worldwide dissemination of research. Founded in 1994, it is now composed of a number of specialized research networks.
- SSRN's email abstract eJournals cover over 1,000 different subject areas
- the Abstract Database contains information on over 756,400 scholarly working papers and forthcoming papers
- the eLibrary currently contains over 639,200 downloadable electronic documents
To enable SSRN as an online import source please visit Administrator > Research output > Import sources and select SSRN. SSRN can be enabled right away; no subscription is required.
Please note that keywords can only be imported from SSRN if you have user-defined keywords enabled on Research Outputs. User-defined keywords are by default enabled, so unless you have edited this setting, you should not be impacted by this feature. If you are unsure, please contact the Pure Support Team.
Additionally, as SSRN also provides DOIs and links, with information on the Public access to file of these electronic versions, we have updated our Open Access : Logic for OA flag document to outline the logic employed
Open Access: Update to Embargo lifting job
We have always taken a conservative approach and lifted the embargo on full text files on the day after the embargo date entered in Pure. In this release, we have adjusted this to be consistent with how repository systems handle the embargo end date and Pure now lifts the embargo on the end date.
5.10
Research Outputs : New unified model
In 5.10 we have unified the Research output datamodels (previously called 'Publications' for some institutions). This means that the underlying metadata model is now aligned across all users of Pure, enabling us to consolidate and future-proof the code base of Pure. Additionally, we have included new functionality:
- More flexibility for configuring the Research output content type to your use cases
- Expanded bulk-edit functionality for Research output
- Alignment of functionality and templates across all Pure users
As, different metadata models existed across the Pure code bases prior to this release, the consequences of this unification will be different according to the code-base of your Pure installation. For the majority of Pure clients, the changes to the Research output metadata model is minimal and you will only notice the new functionality accompanying the unification. Consequences per code-base can be found Research Output Model Details .
Flexible configuration of Research output types
You can now configure Research output types in the similar way as was introduced for Activities, Prizes, Press/Media and other content types with the new Activities data model in the 5.7.0 release.
With this new feature you can now add Research output sub-types specific to your institution. You can also delete sub-types where there are no associated existing records. (Note that some sub-types cannot be deleted as they are used in import sources, but for many of these, you can disable them if they are not relevant for your institution).
Pure Administrators can configure templates and types on the Administrator tab > Research output > Templates and types then by clicking Edit on a particular template.
In these screens, you can define the Categories and peer review relation options available for each sub-type (such as whether records can be marked as peer reviewed, or international peer reviewed) You can delete a Research output sub-type (where possible) by clicking the (-) icon next to the type. Remember to Save the tab afterwards.
Items to note:
- A few Research output sub-types are so important for our import from external sources functionality that the can neither be disabled nor deleted. For example, the type Article in the Contribution to journal template.
- Please be aware that changing Research output sub-types will have consequences when importing and exporting Research Output,and when integrating with external entities (e.g. ORCID, ERA, SEP / KUOZ) due to any mappings defined.
- Research output sub-types can only be deleted if there is no associated content (see section below)
Bulk change of Research output sub-types
Research output templates and sub-types can only be disabled if there is no associated content, i.e. no Research outputs assigned to that template or sub-type. With this in mind, we have introduced new Bulk Edit functionality that enables you to bulk change Research output templates and sub-types.
| To bulk edit the Research output sub-type | |
|---|---|
| 1. | In the Research output editable list, filter for those Research output records with the sub-type you want to delete. |
| 2. | Select all records (or the just the records you want to bulk edit the sub-type of) 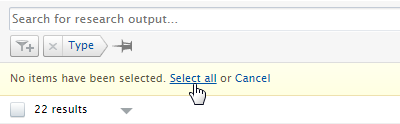
|
| 3. | Select the Bulk edit option that appears.
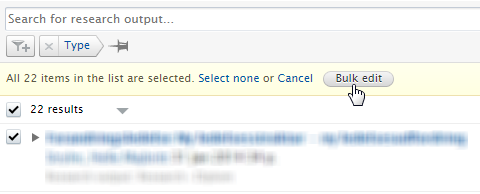
|
| 4. | Select the Change template option and choose the new template and type for the records.
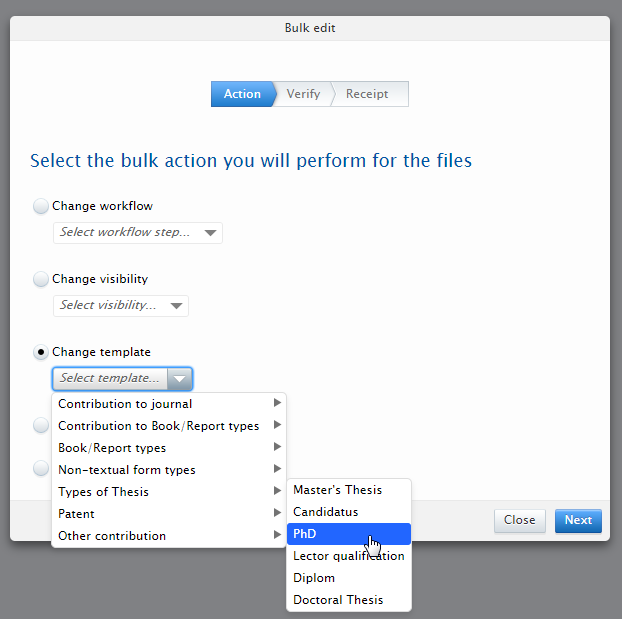
|
| 5. | Define values so that the result so that any new values between the old and new types have a mapping
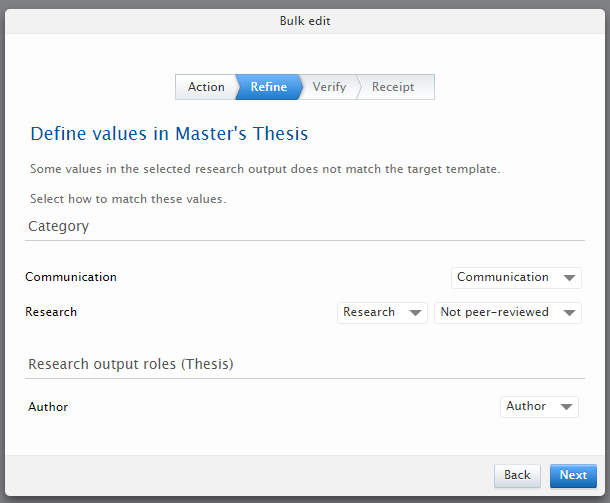
|
| 6. | Follow the remaining steps in the wizard. At the conclusion of the wizard you can download an XLS file with the changes that were made during this bulk operation. |
'Note' functionality (i.e. configurable Description fields) added for all Research output templates
Pure Administrators can now configure custom note fields (i.e. configurable Description fields) for Research output types via the Administrator tab > Research output > Templates and types, enabling greater flexibility in ensuring that the Research Output template meets your data needs.
Open Access improvements
In this release, we have expanded the "Allow Personal Users / Editor of Research Output to delete electronic versions (files, DOIs, and links) on approved / validated research output records" configurations to also include a restriction on Personal Users / Editor of Research Output being able to delete / edit publication states and dates on validated / approved records. This will help to ensure that the full publication chronology is maintained as the Research Output metadata record evolves during the publication process.
In 5.8.0, we introduced new configurations that restricted Personal users and Editors of Research Outputs from being able to delete electronic versions (files, DOIs, and links) on approved / validated research output records. Now, in 5.10, we have expanded these configurations to also include a restriction on Personal users / Editors of Research Outputs being able to delete / edit existing publication states and dates on validated / approved records. For example, if the validated record has an Accepted date input, the Personal user will not be able to edit / delete this state, only add additional states and dates.
The configuration is available in Administrator > System settings > Publication - Allow Personal Users / Editor of Research output to delete electronic versions (files, DOIs, and links) and update publication statuses on approved / validated research output records.
5.9
Open Access Improvements - Pure portal
This release includes a number of improvements to aid visitor discovery of Open Access publications within the Pure Portal. This includes the display of icons to indicate Open Access status and the presence of an attached document, and the ability to filter for Open Access content. We are also providing the license information as a hyperlink, and the Bibliographical note field is now available for display in the Portal too, enabling customers to show publisher rights statements as required.
5.8
Open Access Improvements
This release includes:
- Expansion of the Journal datamodel to include whether the Journal is indexed in Directory of Open Access Journals (DOAJ)
- New Unknown value available for 'Public access to file'
- Improvements to functionality relating to previously embargoed electronic versions
Improvements to restrictions on users from deleting / replacing Electronic versions from validated records
The Directory of Open Access Journals (DOAJ) "is a community-curated online directory that indexes and provides access to high quality, open access, peer-reviewed journals."
We have expanded the Pure Journal datamodel to include whether the Journal is indexed in DOAJ, enabling users to identify outputs published in Journals indexed in DOAJ. This DOAJ information appears in the Journal editor, in addition to renders of the Journal (on, for example, the Research Output and Activity editors). In the Reporting module, it is possible to filter, group, and report on Journals indexed in DOAJ (both when reporting on Journals by themselves, or as a related content type).
We have improved two elements relating to the Open Access Status of previously embargoed electronic versions:
1. Automatic updating of the Open Access Status once the Embargo End Date has passed
In recent testing of the OA Status functionality, and as per customer feedback, it was determined that the existing functionality that was designed to update the Open Access Status of outputs once the embargo period had passed (as described in the Open Access : Logic for OA Flag (PDF) documentation) was not functioning as expected. As such, we have introduced a new "Open Access Embargo Lifted Job" that finds those outputs where the Embargo End Date is the previous day (e.g. if the job runs at 00.30 on 1 December, it checks all those where the embargo end date is 30 November) and changes (where appropriate) the Open Access Status from 'Embargoed' to 'Open'.
This job is enabled by default, and is scheduled to run nightly just after midnight.
2. Improvements to Filters (in Reports and Editable lists)
In order to support more accurate OA reporting and monitoring, we have updated the 'Open Access Status' filters such that content where the Open Access Status was previously 'Embargoed' will now be included in the 'Open Access Status' = Open filter, thereby reflecting their actual current state in terms of accessibility. Should you wish to also see those outputs that had been embargoed in the past, you can filter the list / report further by using the Period > Open Access Embargo date filter.
Individual electronic versions that had been under embargo will remain with the 'Public access to file' of Embargoed in order to continue to facilitate detailed reporting on the metadata of each individual electronic version.
Published at June 10, 2024
